Physiological and metabolomic analysis of Plantago asiatica L. in response to cadmium stress
-
摘要:
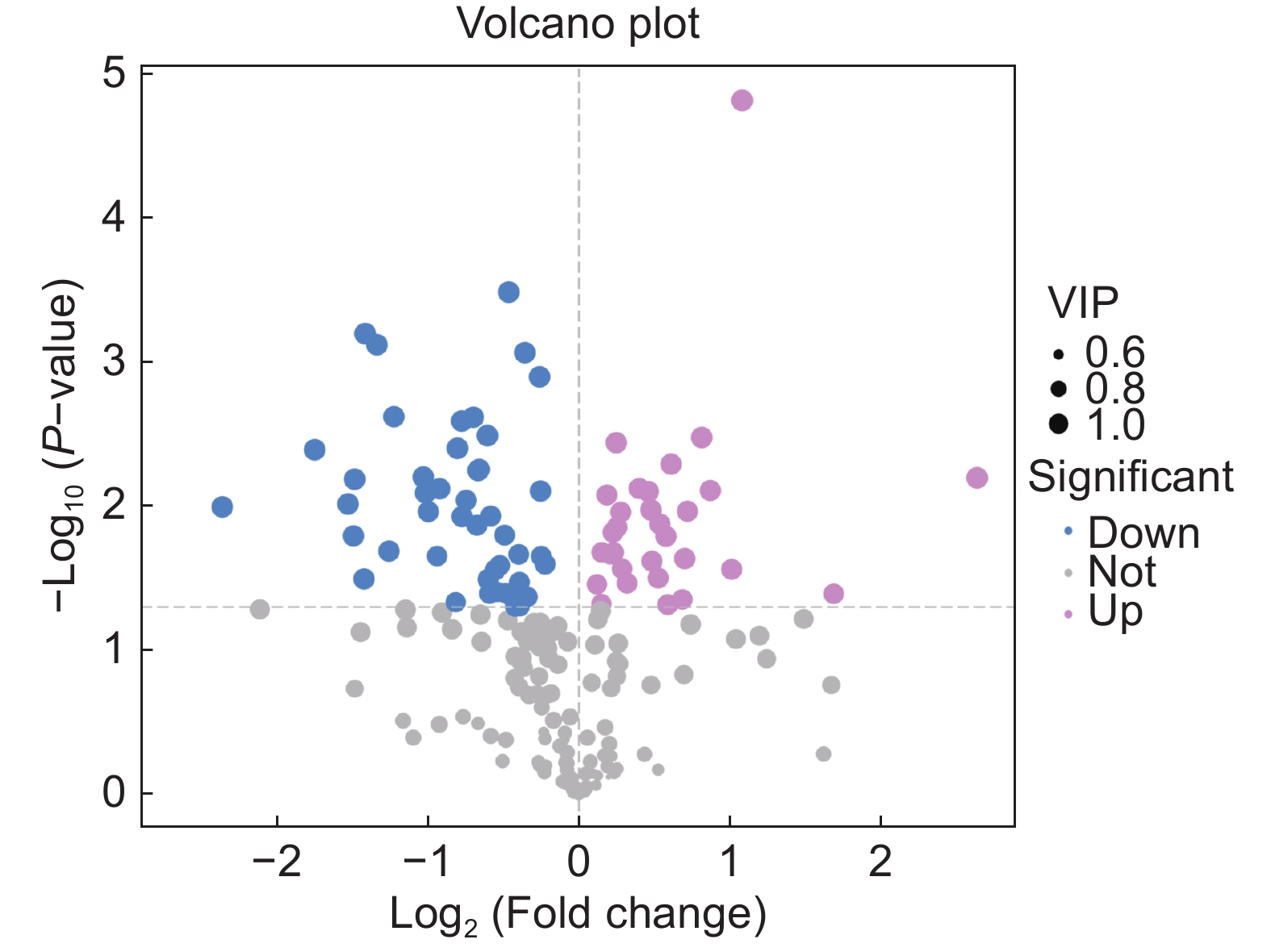
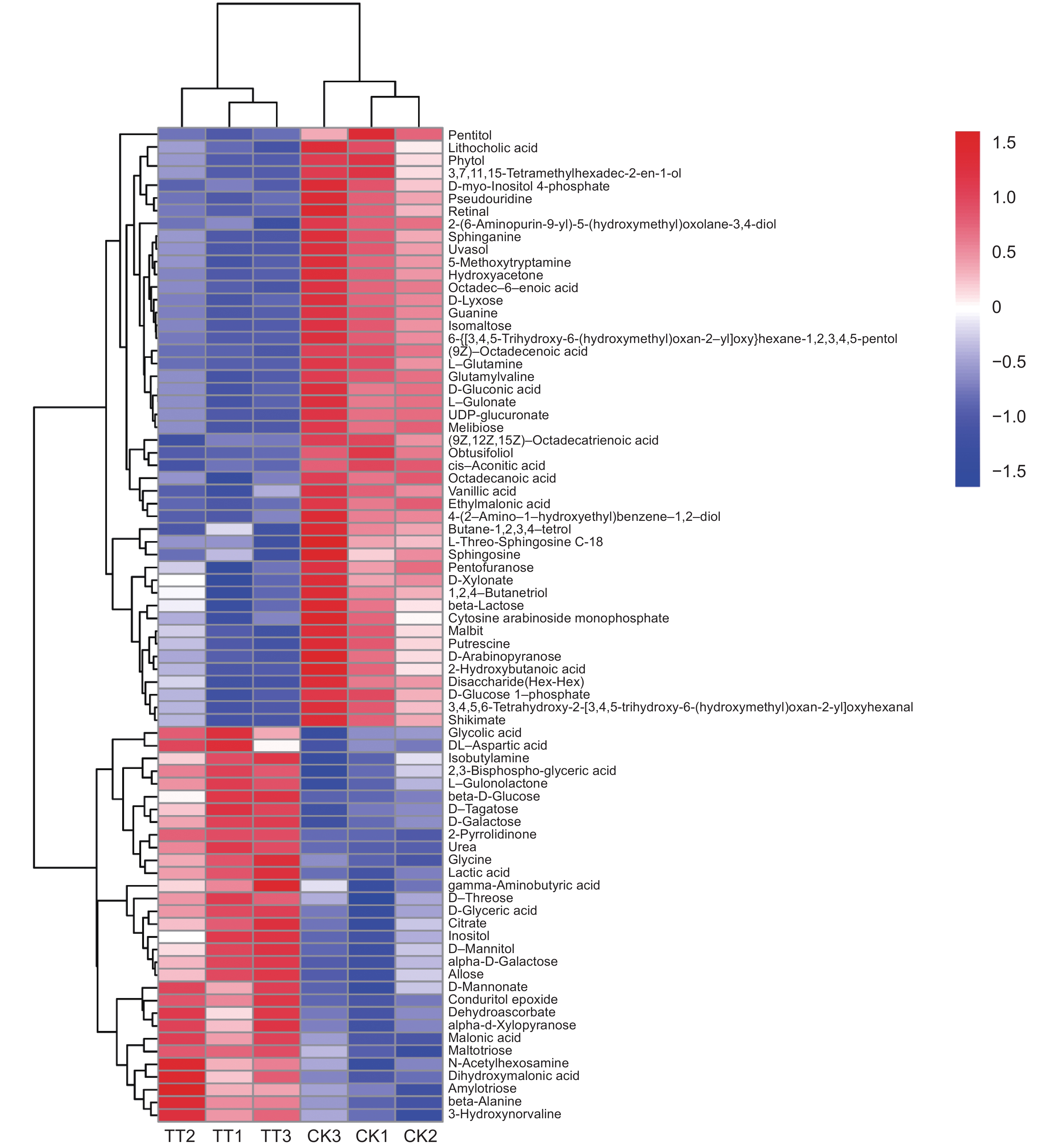
本研究测定了镉(Cd)胁迫下车前(Plantago asiatica L.)超氧化物歧化酶(SOD)、过氧化物酶(POD)、过氧化氢酶(CAT)的活性以及还原型谷胱甘肽(GSH)的含量,以探明车前响应Cd胁迫的生理机制,并进一步利用代谢组学方法筛选差异代谢物,并进行代谢通路富集分析。结果显示,随着Cd浓度的升高,SOD和CAT活性显著下降,POD活性显著上升,且总体呈先上升后下降的趋势,而GSH含量没有显著变化。50 mg/kg Cd胁迫组(YT3)与对照组(YT0)之间的显著差异代谢物有78个,其中31个上调,47个下调,且差异代谢物主要集中在糖类和氨基酸类。差异代谢物主要富集到20条通路中,包括3条糖代谢途径和3条脂类代谢途径。
Abstract:In this study, the activities of superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) and content of reduced glutathione (GSH) in Plantago asiatica L. under cadmium (Cd) stress were determined to explore the biological mechanisms of P. asiatica in response to such stress. Metabolomics was further used to screen differential metabolites, and metabolic pathway enrichment analysis was conducted. Results showed that with the increase in Cd concentration, the activities of SOD and CAT decreased significantly, POD activity increased significantly, with an overall trend of first increasing and then decreasing. GSH content showed no significant differences. There were 78 metabolites showing significant differences between the 50 mg/kg Cd stress group (YT3) and control group (YT0), including 31 up-regulated and 47 down-regulated metabolites, mainly carbohydrates and carboxylic acids. The differential metabolites were mainly enriched in 20 pathways, including three glucose metabolism pathways and three lipid metabolism pathways.
-
Keywords:
- Plantago asiatica /
- Cd stress /
- Physiological indexes /
- Metabonomics
-
地黄(Rehmannia glutinosa Libosch.)为玄参科地黄属植物,多年生草本,以块根入药,为著名的“四大怀药”之一。地黄最早记载见《神农本草经》,被列为上品,迄今已有2000余年的应用历史。根据炮制方法的不同,地黄药材分为鲜地黄、生地黄和熟地黄[1]。2020版《中国药典》记载,鲜地黄具有清热生津、凉血、止血的功效,生地黄具有清热凉血、养阴生津的功效,熟地黄具有补血滋阴,益精填髓的功效[2]。地黄富含环烯醚萜类、苯乙醇苷类、紫罗兰酮类、三萜类、黄酮类和糖类等,对人体心脑血管、血液、中枢神经和免疫系统等均有显著作用[3]。
毛蕊花糖苷(Acteoside)是地黄中含量较高的苯乙醇苷类化合物[4],具有抗氧化、免疫调节、抗炎、保肝、抗肿瘤、增强记忆力等生物活性[5],是2010版、2015版《中国药典》规定的地黄药材质量控制的指标性成分之一。地黄毛蕊花糖苷的含量易受品种[6]、产地[7]、收获时期[5]、种植密度[8]和光照条件[9]等因素的影响,造成某些年份部分地黄药材的毛蕊花糖苷含量达不到《中国药典》规定的要求。地黄毛状根中也含有丰富的毛蕊花糖苷,本课题组前期研究表明,在毛状根诱导的特定时期添加水杨酸(SA)可显著促进毛蕊花糖苷的含量[10]。然而,地黄生长发育过程中叶面喷施SA对毛蕊花糖苷的含量是否有影响还未见报道。
本研究以大田栽培的地黄为材料,采用叶面喷施方法分析SA对地黄叶片和块根毛蕊花糖苷含量的影响,并利用转录组测序技术分析地黄块根中的基因表达特征,研究结果旨在为生产中应用外源激素提高毛蕊花糖苷的含量提供理论依据。
1. 材料与方法
1.1 实验材料
供试材料为地黄‘温85-5’,种植在河南省武陟县蔡庄村(35°2′51″N,113°18′34″E),经河南农业大学王丰青教授鉴定为Rehmannia glutinosa Libosch.。将浓度为100 μmol/L的SA水溶液均匀喷施在生长180 d处于膨大后期长势一致的地黄叶片上,以叶片完全湿润且无水滴落下为准。分别于处理后1、3和6 h进行取样,取样时选择位置相同的叶片。对照组喷施蒸馏水,与处理组同时取样。样品清洗干净后,将块根切成1 cm3左右的小块,叶片剪碎,55℃烘干后打粉,过3号药典筛。样品粉末存放于干净的8号自封袋中备用。
1.2 毛蕊花糖苷含量测定
对照品毛蕊花糖苷(批号MUST-18032725)购于成都曼斯特科技有限公司,纯度均 ≥ 98%。采用 Agilent1260高效液相色谱仪(美国安捷伦科技有限公司)进行检测。
色谱条件:采用的色谱柱型号为 Dikma Diamonsil C18(4.6 × 250 mm,5 μm),柱温30℃,流速1 min/mL。毛蕊花糖苷的流动相为乙腈−0.1%醋酸水(16 : 84),检测波长334 nm,进样量为20 μL。
供试样品溶液制备:精密称取地黄叶和块根样品粉末0.8 g,放入锥形瓶中,精密吸取50 mL甲醇加入锥形瓶中,称重,并在65℃加热回流提取1.5 h,放凉至室温称重,用甲醇补足失重后,摇匀过滤。于蒸发皿中精密吸取滤液20 mL进行毛蕊花糖苷分析,在电热恒温水浴锅上浓缩至近干,残渣用流动相溶解,转移至5 mL容量瓶中,用流动相稀释至刻度,摇晃均匀,用0.22 μm微孔的滤膜过滤,滤液装入2 mL的进样瓶待测。
含量计算:以本实验室建立的标准曲线Y = 30024X − 110.9来计算毛蕊花糖苷含量,Y为峰面积积分值,X为样品的质量浓度。
1.3 样品RNA提取及高通量测序
用TRIzol试剂提取样品的总RNA,用核酸测定仪检测RNA的浓度和质量。使用带有Oligo dT的磁珠富集具有polyA尾巴的mRNA,然后将RNA片段化,反转录后再合成cDNA第2链,形成双链cDNA。双链cDNA经过末端修复、3′末端加A、添加测序接头、多轮扩增、热变性成单链及单链环化等一系列步骤后,完成测序文库的制备。测序委托华大基因科技有限公司进行,测序平台为BGISEQ-500。
对测序得到的原始序列(Raw reads)进行质控处理,去除低质量、接头及污染序列,获取过滤后的测序序列(Clean reads)。使用Bowtie2将Clean reads比对到课题组前期获得的地黄叶和根的参考基因序列集[11],统计不同样品的片段序列比对率以及分布,之后再使用RSEM 计算基因的表达水平,表达量用FPKM表示。对比分析水杨酸喷施前后地黄块根中基因的表达水平,获取水杨酸喷施处理的特异响应基因,并对其进行GO注释和KEGG注释,根据注释结果进行功能分类和KEGG pathway分类,并使用R软件中的phyper函数进行富集分析,获得水杨酸喷施处理下地黄块根内的关键分子响应进程。
1.4 实时荧光定量(qRT-PCR)分析
用TaKaRa反转录试剂盒对RNA进行反转录,合成cDNA,反应体系包括1 μL oligo dT primer,1 μL dNTP Mixture,2 μg 模板RNA,加水补足体积到10 μL,65℃保温5 min后,冰上迅速冷却。再加0.5 μL的RNase抑制剂,1 μL PrimeScript Ⅱ RTase,4 μL 5 × PrimeScript Ⅱ Buffer,加水补足体积到20 μL。反应程序为42℃ 60 min,95℃ 5 min。以RgTIP41为内参基因,用实时荧光定量PCR检测基因表达水平。所用试剂盒为SYBR® Premix Ex Taq™ Ⅱ(Tli RNaseH Plus) (Takara,大连),使用仪器为Bio-Rad IQ5(上海伯乐公司)。定量反应体系为25 μL,包含2 μL 上述反转录cDNA产物,上、下游引物各1 μL,12.5 μL SYBR® Premix Ex Taq,8.5 μL ddH2O。反应程序为:95℃变性30 s;然后95℃ 5 s, 60℃ 30 s,40个循环。结束反应后获得不同样品的扩增循环数Ct,使用2-ΔΔCt法计算不同基因的相对表达量。
2. 结果与分析
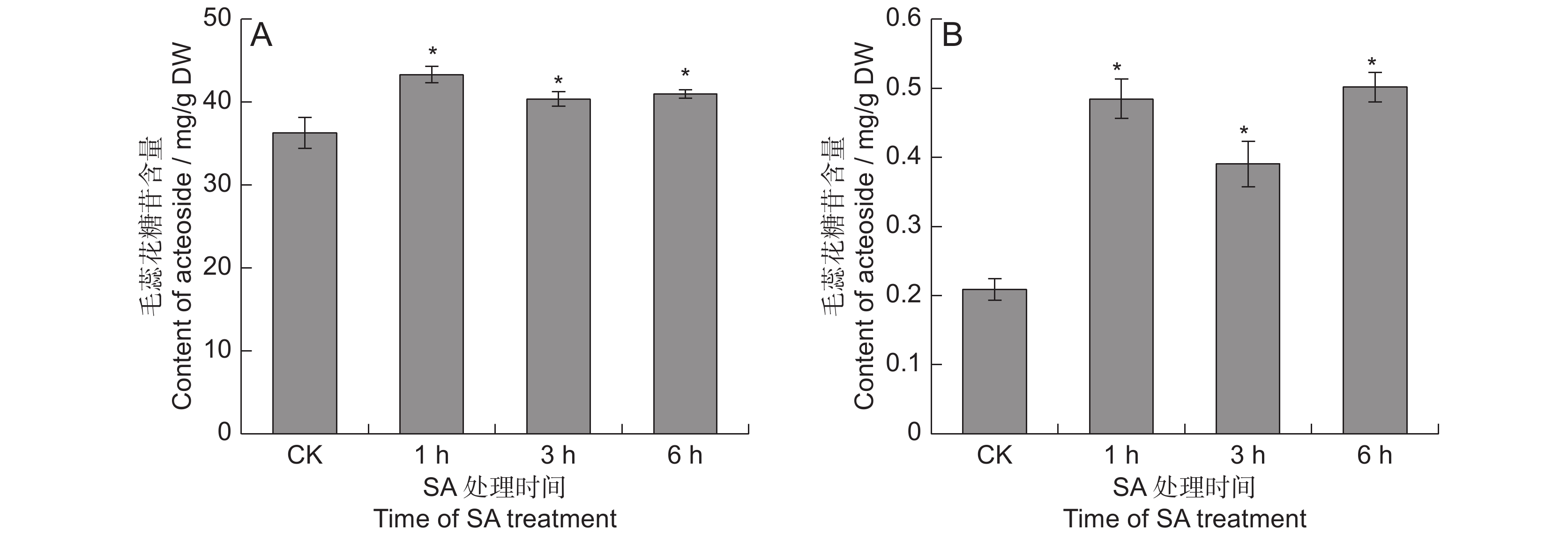
2.1 SA处理对地黄毛蕊花糖苷含量的影响
对SA处理的地黄叶和块根中的毛蕊花糖苷含量进行测定,结果表明,SA能够显著提高毛蕊花糖苷的含量(图1)。在叶中, SA处理1 ~ 6 h后,毛蕊花糖苷含量分别提高了11.2% ~ 19.3%。块根中毛蕊花糖苷的提升幅度远高于叶片,分别较对照提高了0.9 ~ 1.4倍,处理 6 h后的毛蕊花糖苷含量最高,达0.5 mg/g,远超2015版《中国药典》规定的0.02%,说明地黄叶面喷施SA可显著提高叶和块根中毛蕊花糖苷的含量。
2.2 地黄块根RNA测序分析
利用Agilent 2100 Bioanalyzer和Fragment Analyzer分别对提取的各样品总RNA质量进行检测,结果显示,12个样品的总RNA浓度在430 ~ 1260 ng/μL,总RNA质量在8.6 ~ 25.2 μg,RNA的浓度和总量满足建库需求。RIN值在8.2 ~ 9.9,28S/18S > 1.6,说明RNA较为完整,符合建库要求。
为了分析SA处理后地黄块根相关基因的表达特性,对SA处理1、3和6 h后的地黄块根进行RNA-seq分析,结果表明(表1),每个测序样本获得的总原始读段量均为21.94 M,去除低质量、接头污染及未知碱基N含量过高的reads,获得的高质量reads在20.98 ~ 21.27 M,碱基数均在1.05 ~ 1.06 Gb,测序数据量基本一致。将每个样品的测序数据匹配地黄参考转录组,发现匹配率在85.04% ~ 87.70%,特异匹配率在47.35% ~ 51.31%,测序数据能够较好地反映细胞中基因表达的真实情况,说明测序质量良好,可以进行后续基因表达分析。
表 1 测序数据统计结果Table 1. Statistics of sequenced data样本
Sample总原始序列
Total raw reads / M总测序序列
Total clean reads / M总测序碱基数
Total clean bases / Gb测序序列比率
Clean read ratio / %总匹配率
Total mapped / %特异匹配率
Uniquely mapped / %Control_1 21.94 21.13 1.06 96.3 87.70 51.31 Control_2 21.94 21.07 1.05 96.04 86.92 51.18 Control_3 21.94 21.08 1.05 96.08 85.72 51.09 SA 1h_1 21.94 21.27 1.06 96.93 85.44 47.35 SA 1h_2 21.94 21.08 1.05 96.05 87.07 48.03 SA 1h_3 21.94 21.15 1.06 96.37 87.28 49.26 SA 3h_1 21.94 20.98 1.05 95.59 86.85 50.36 SA 3h_2 21.94 21.05 1.05 95.93 85.74 49.92 SA 3h_3 21.94 21.05 1.05 95.95 86.81 50.47 SA 6h_1 21.94 21.09 1.05 96.11 86.50 50.25 SA 6h_2 21.94 21.06 1.05 96.00 85.04 49.42 SA6h_3 21.94 21.03 1.05 95.86 86.68 49.97 2.3 SA处理前后差异表达基因分析及筛选
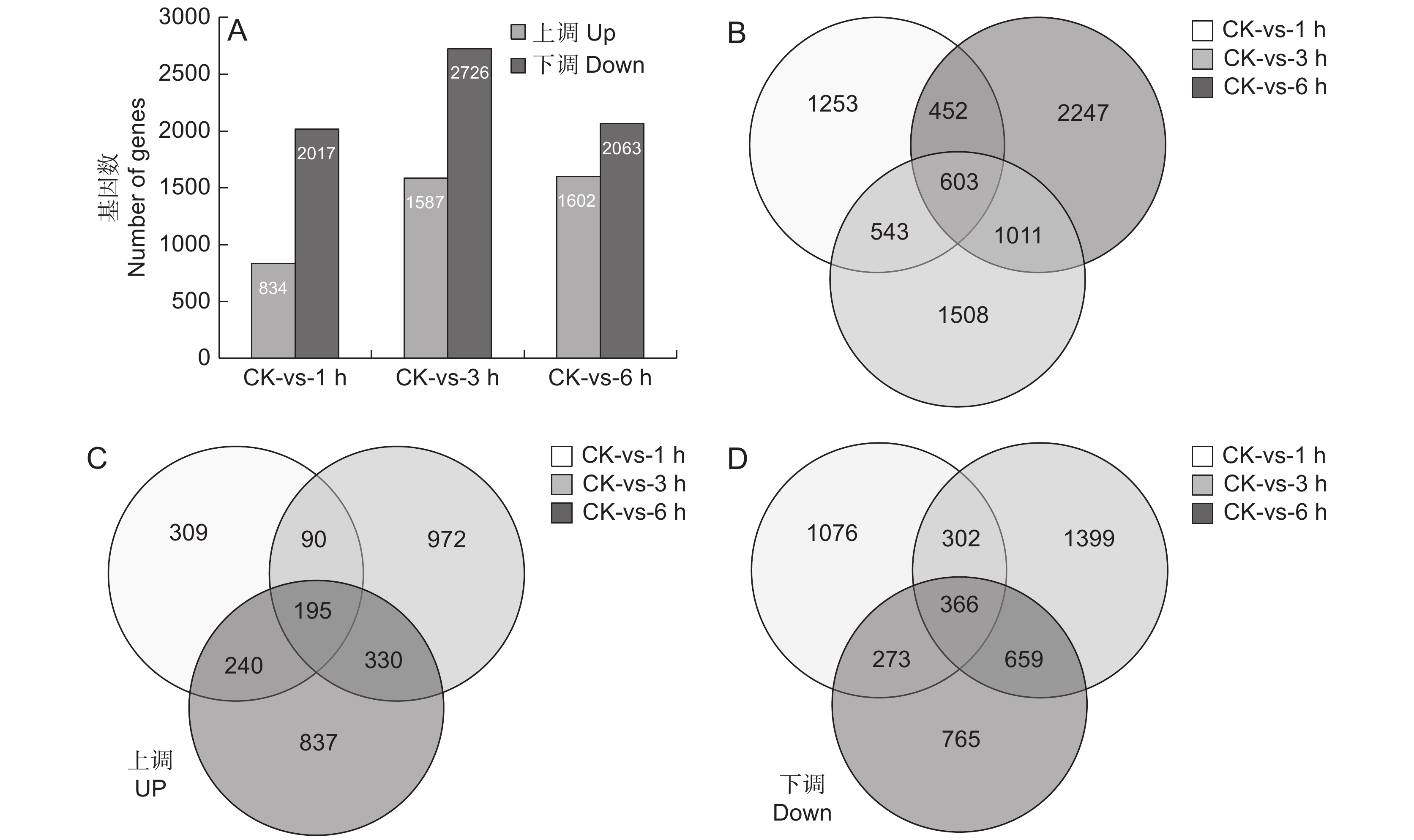
为了分析SA处理后块根中基因的表达特性,采用FPKM值比较基因丰富度的差异(图2)。使用以下标准对差异表达基因(DEGs)进行识别和筛选:校正P值 < 0.001且log2值 ≥ 2。分析SA处理不同时间后与对照样品中的差异表达基因(图2:A),发现SA处理1 h后834个基因上调,2017个基因下调;处理3 h后1587个基因上调,2726个基因下调;处理6 h后1602个基因上调,2063个基因下调。进一步分析SA处理后的3个时间点与CK相比的共同差异表达基因,发现共有603个基因是共同差异表达的(图2:B),其中上调表达和下调表达的基因数分别为195(图2:C)和366个(图2:D)。
![]() 图 2 SA处理后基因显著差异表达A:SA处理过程中上调和下调的基因数目; B ~ D:SA处理后不同时间点鉴别出的总DGEs(B)、上调DGEs(C)和下调DGEs的维恩图。Figure 2. Significant DEGs in response to SA treatmentA: Up-regulated and down-regulated gene numbers during SA treatment; B − D: Venn diagram of total DEGs (B), up-regulated DEGs (C), and down-regulated DEGs (D) identified at different time points after SA treatment.
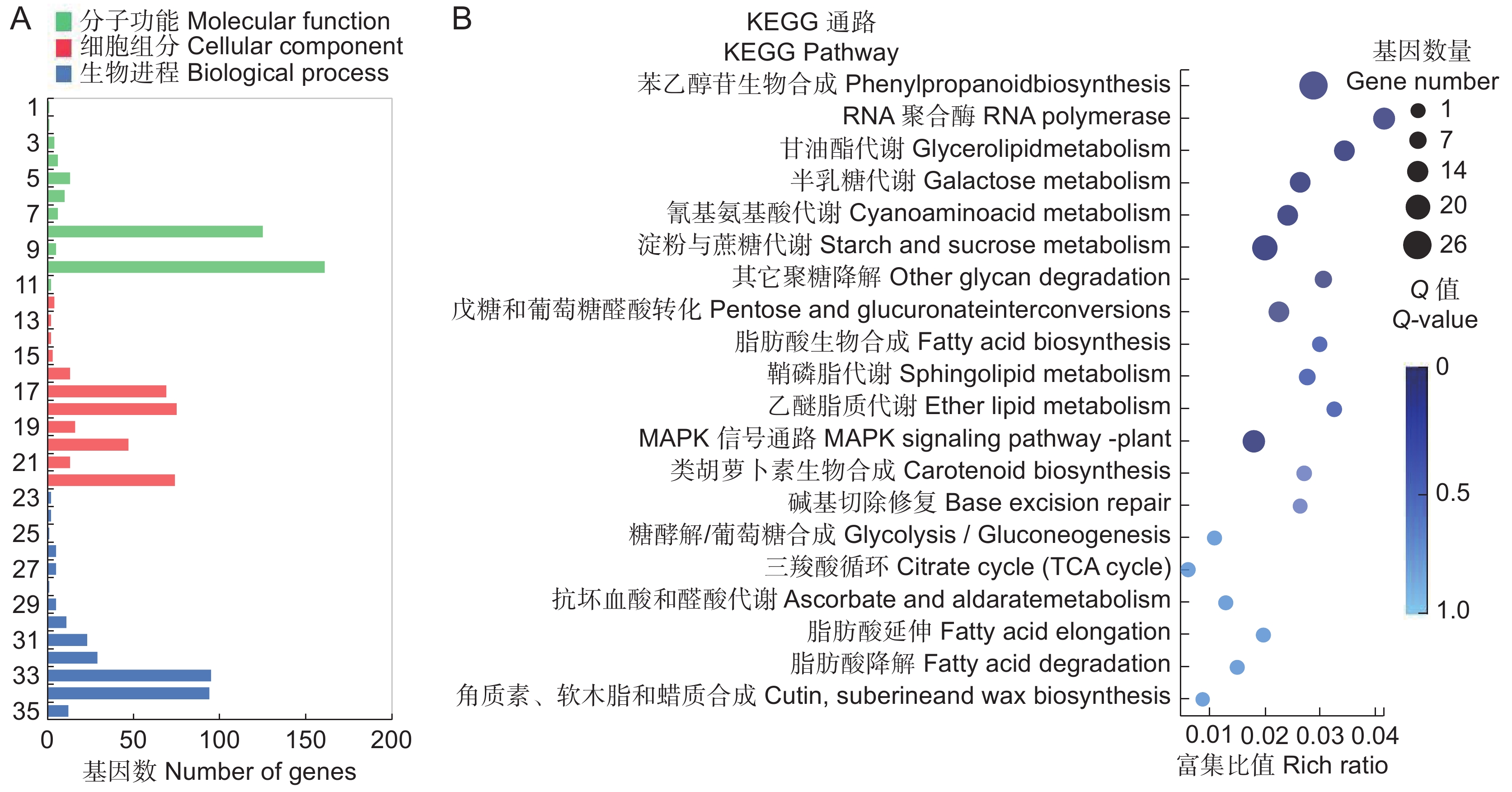
图 2 SA处理后基因显著差异表达A:SA处理过程中上调和下调的基因数目; B ~ D:SA处理后不同时间点鉴别出的总DGEs(B)、上调DGEs(C)和下调DGEs的维恩图。Figure 2. Significant DEGs in response to SA treatmentA: Up-regulated and down-regulated gene numbers during SA treatment; B − D: Venn diagram of total DEGs (B), up-regulated DEGs (C), and down-regulated DEGs (D) identified at different time points after SA treatment.对603个DGEs进行GO功能分类(图3:A),结果显示其共分为分子功能(Molecular function)、细胞功能(Cellar function)和生物功能(Biological function)3个大类。其中分子功能分类中的催化活性(Catalytic activity)占比最多,其次是ATP结合;在细胞功能分类中,细胞(Cell)、细胞膜(Membrane)、细胞膜构件(Membrane part)较多;生物功能分类中,细胞过程(Cellular process)和代谢过程(Metabolic process)所占比重最多。进一步对603个DEGs进行KEGG代谢通路富集分析,图3:B展示了最显著的前20个代谢通路,这些通路涉及各项生命活动。其中首先被富集的是苯乙醇苷生物合成通路(Phenylpropanoid biosynthesis),其次是淀粉和蔗糖代谢合成通路(Starch and sucrose metabolism)、植物MARK信号通路(MAPK signaling pathway plant)及RNA聚合酶通路(RNA polymerase)。这说明SA喷施对地黄块根中次生代谢物的积累产生了较大影响,且调控了碳水化合物和MAPK等多个代谢通路。
![]() 图 3 SA处理后3种比对均差异表达基因的GO分类及KEGG通路富集A:603个基因的GO分类;B:KEGG富集的前20个代谢通路。圆点大小和颜色分别表示通路中DEGs的数量和Q值范围。1:分子转导活性;2:分子载体活性;3:信号转导活性;4:结构分子活性;5:转运活性;6:转录调节活性;7:抗氧化活性;8:结合;9:分子功能调节;10:催化活性;11:膜封闭腔;12:超分子复合物;13:共质体;14:细胞连接;15:细胞组分;16:细胞外区域;17:膜组分;18:膜;19:细胞器组分;20:细胞器;21:大分子复合物;22:细胞;23:生殖过程;24:繁殖;25:解毒作用;26:多细胞生物过程;27:发育过程;28:多生物体过程;29:信号;30:定域化;31:生物调节;32:对刺激的反应;33:代谢过程;34:细胞过程;35:细胞成分组织或生物合成。Figure 3. GO classification and KEGG pathway enrichment of co-DEGs in three comparisonsA: GO classification of 603 genes; B: Top 20 enriched KEGG pathways among 603 genes. Size and color of dot represent number and scope of DEGs in pathway, respectively. 1: Molecular transducer activity; 2: Molecular carrier activity; 3: Signal transducer activity; 4: Structural molecule activity; 5: Transporter activity; 6: Transcription regulator activity; 7: Antioxidant activity; 8: Binding; 9: Molecular function regulator; 10: Catalytic activity; 11: Membrane-enclosed lumen; 12: Supramolecular complex; 13: Symplast; 14: Cell junction; 15: Cell part; 16: Extracellular region; 17: Membrane part; 18: Membrane; 19: Organelle part; 20: Organelle; 21: Macromolecular complex; 22: Cell; 23: Reproductive process; 24: Reproduction; 25: Detoxification; 26: Multicellular organismal process; 27: Developmental process; 28: Multi-organism process; 29: Signaling; 30: Localization; 31: Biological regulation; 32: Response to stimulus; 33: Metabolic process; 34: Cellular process; 35: Cellular component organization or biogenesis.
图 3 SA处理后3种比对均差异表达基因的GO分类及KEGG通路富集A:603个基因的GO分类;B:KEGG富集的前20个代谢通路。圆点大小和颜色分别表示通路中DEGs的数量和Q值范围。1:分子转导活性;2:分子载体活性;3:信号转导活性;4:结构分子活性;5:转运活性;6:转录调节活性;7:抗氧化活性;8:结合;9:分子功能调节;10:催化活性;11:膜封闭腔;12:超分子复合物;13:共质体;14:细胞连接;15:细胞组分;16:细胞外区域;17:膜组分;18:膜;19:细胞器组分;20:细胞器;21:大分子复合物;22:细胞;23:生殖过程;24:繁殖;25:解毒作用;26:多细胞生物过程;27:发育过程;28:多生物体过程;29:信号;30:定域化;31:生物调节;32:对刺激的反应;33:代谢过程;34:细胞过程;35:细胞成分组织或生物合成。Figure 3. GO classification and KEGG pathway enrichment of co-DEGs in three comparisonsA: GO classification of 603 genes; B: Top 20 enriched KEGG pathways among 603 genes. Size and color of dot represent number and scope of DEGs in pathway, respectively. 1: Molecular transducer activity; 2: Molecular carrier activity; 3: Signal transducer activity; 4: Structural molecule activity; 5: Transporter activity; 6: Transcription regulator activity; 7: Antioxidant activity; 8: Binding; 9: Molecular function regulator; 10: Catalytic activity; 11: Membrane-enclosed lumen; 12: Supramolecular complex; 13: Symplast; 14: Cell junction; 15: Cell part; 16: Extracellular region; 17: Membrane part; 18: Membrane; 19: Organelle part; 20: Organelle; 21: Macromolecular complex; 22: Cell; 23: Reproductive process; 24: Reproduction; 25: Detoxification; 26: Multicellular organismal process; 27: Developmental process; 28: Multi-organism process; 29: Signaling; 30: Localization; 31: Biological regulation; 32: Response to stimulus; 33: Metabolic process; 34: Cellular process; 35: Cellular component organization or biogenesis.2.4 毛蕊花糖苷合成相关催化酶基因表达分析
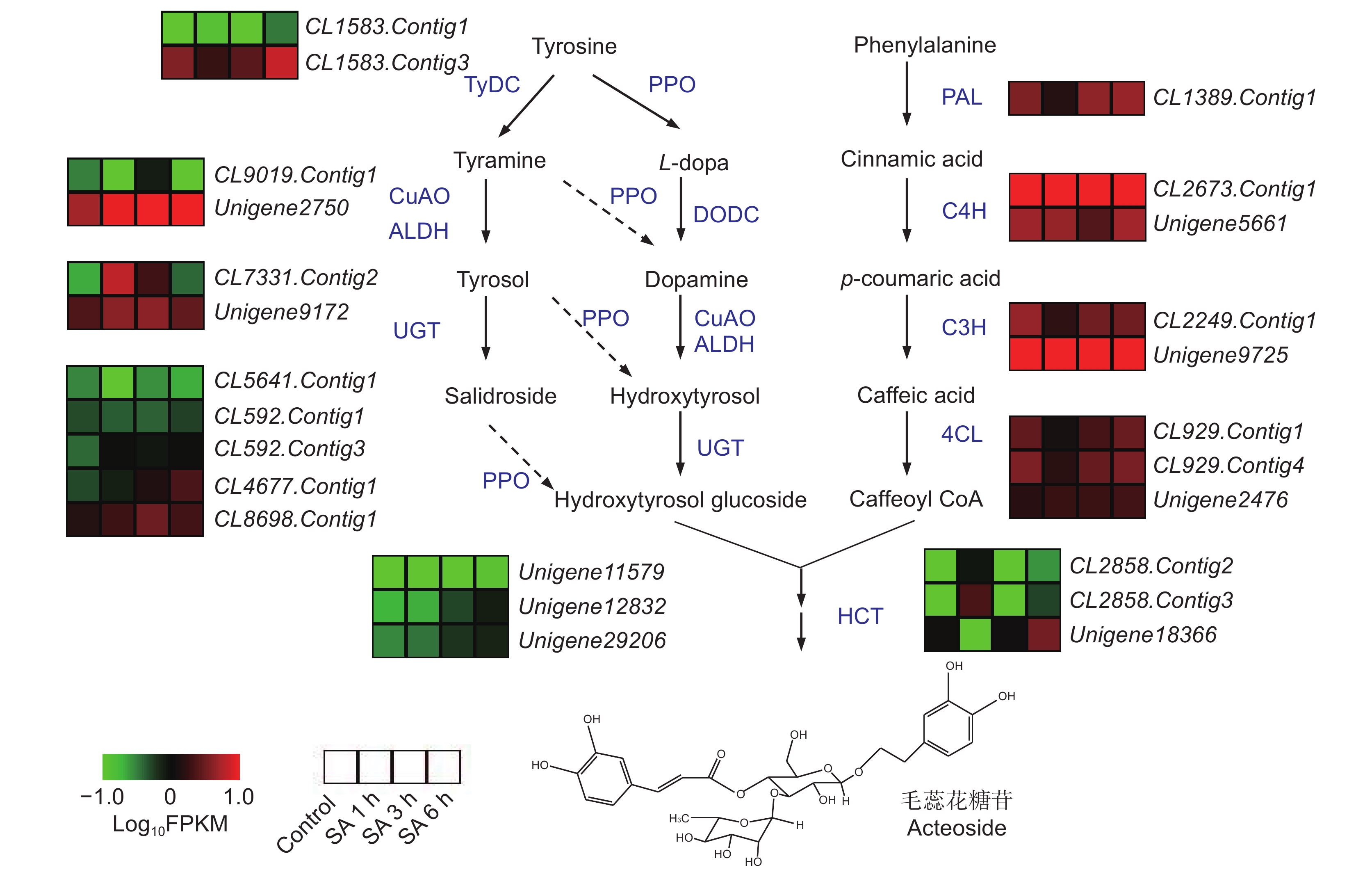
研究表明,在植物体内,毛蕊花糖苷由苯丙氨酸途径的咖啡酰辅酶A(Caffeoyl CoA)和酪氨酸途径的羟基酪醇苷(Hydroxtrosol glucoside)经缩合糖苷化后生成[8]。在地黄转录组中鉴定出可能参与毛蕊花糖苷合成的编码催化酶的基因215个,但SA处理后,地黄块根中仅有少数催化酶基因上调表达(图4)。在酪氨酸途径中,编码乙醛脱氢酶(ALDH)的基因CL7331.Contig2,在SA处理1 h和3 h后的地黄块根中表达量显著升高,另一个ALDH编码基因Unigene9172在SA处理后的表达量也有一定程度上升。编码糖苷转移酶(UGT)的基因CL4677.Contig1随着SA处理时间的延长其表达量逐渐升高,在SA处理6 h后表达量最高。编码多酚氧化酶(PPO)的两个基因Unigene12832和Unigene12832在SA处理3 h和6 h后的表达量增加较为明显。而苯丙氨酸途径的催化酶基因表达变化不明显。这说明SA处理后毛蕊花糖苷含量的增加可能主要与酪氨酸途径部分催化酶基因的表达量增加有关。
2.5 差异表达转录因子筛选
对SA处理不同时间后地黄块根中差异表达的转录因子进行分析,结果表明(表2),25种转录因子的编码基因在处理前后呈差异表达。在处理1和3 h后,下调表达的转录因子较多,处理6 h后则上调表达的转录因子较多。WRKY、MYB、bHLH、AP2-EREBP、NAC和GRAS转录因子的差异表达基因较多,其中AP2-EREBP、WRKY和MYB的差异表达基因最多,且均表现为SA处理1 h后下调的基因较多, 3 h和6 h后上调表达的基因较多。进一步分析发现,共有20个转录因子编码基因在SA处理后的3个时间点均上调表达(表3),其中NAC和AP2-EREBP基因均为4个,WRKY基因有3个,MYB、GRAS、PLATZ基因各2个,bHLH、MADS和C2C2-CO-lik基因各1个。具有调控毛蕊花糖苷合成功能的RgWRKY37编码基因CL394.Contig2在SA处理1、3、6 h后的Log2(SA处理/CK)的值分别为0.79、0.82和0.57,Log2(SA处理/CK)的值虽然小于1,但其Q-value和P-value均达到了显著水平,说明在SA处理后CL394.Contig2上调表达。
表 2 SA处理后差异表达的转录因子数Table 2. Number of differentially expressed transcription factors (TFs) after SA treatment转录因子
Transcription factorCK-vs-1 h CK-vs-3 h CK-vs-6 h 共同差异表达的基因数
Number of common DEGs下调 Down 上调 Up 下调 Down 上调 Up 下调 Down 上调 Up zf-HD 2 1 1 1 0 0 0 WRKY 11 5 8 16 5 15 5 TUB 1 0 1 1 0 0 0 Trihelix 1 0 1 0 0 0 0 Tify 1 0 3 0 3 0 1 PLATZ 0 4 0 4 1 3 2 SRS 0 0 2 0 2 0 0 OFP 2 0 3 0 1 0 0 NAC 2 8 2 6 0 8 4 MYB 15 10 5 7 6 12 3 mTERF 1 0 2 0 1 1 0 MADS 2 2 3 4 2 2 1 LOB 1 0 4 2 2 0 0 HSF 5 2 4 6 3 6 2 GRAS 1 8 0 4 0 7 2 G2-like 2 0 2 1 0 3 0 CPP 2 0 2 0 4 0 2 C2H2 3 1 8 1 3 1 2 C2C2-GATA 2 0 1 0 5 0 0 C2C2-Dof 1 1 3 0 2 1 0 C2C2-CO-like 0 1 0 2 4 3 1 bZIP 1 2 0 0 1 0 0 bHLH 12 3 15 4 5 4 2 AP2-EREBP 15 12 12 18 9 21 6 ABI3VP1 5 2 7 1 1 1 0 总数 88 62 89 78 60 88 33 表 3 SA处理后上调表达的转录因子基因Table 3. Up-regulated transcription factor genes after SA treatment转录因子
Transcription factor基因
GeneLog2(SA/CK) 功能
Function1 h 3 h 6 h AP2-EREBP CL1637.Contig3 2.82 1.29 2.42 Ethylene-responsive transcription factor ERF071 AP2-EREBP CL4501.Contig2 1.66 1.75 2.16 Pathogenesis-related genes transcriptional activator PTI6 AP2-EREBP CL7827.Contig1 1.77 3.51 4.24 Ethylene-responsive transcription factor ERF106-like AP2-EREBP Unigene2558 1.62 1.06 1.74 Ethylene-responsive transcription factor 2-like MYB CL1983.Contig1 4.55 4.19 3.12 Transcription factor TFIIIB component B''-like MYB CL4303.Contig1 3.76 2.61 3.47 Single MYB histone protein NAC CL2945.Contig2 6.82 7.15 7.51 NAC domain-containing protein 82-like isoform X1 NAC CL4851.Contig1 3.15 1.99 2.44 NAC transcription factor 29 NAC CL4851.Contig2 3.15 2.27 2.82 NAC transcription factor 29 NAC Unigene5193 2.09 1.38 2.37 NAC domain-containing protein 72 C2C2-CO-like CL5505.Contig3 2.94 2.66 4.31 Zinc finger protein CONSTANS-LIKE 4-like PLATZ CL5569.Contig1 2.31 1.49 2.53 Interleukin-1 receptor-associated kinase 4 PLATZ CL5569.Contig3 2.47 1.71 2.38 Interleukin-1 receptor-associated kinase 4 GRAS CL645.Contig2 1.93 1.65 2.91 Scarecrow-like protein 14 GRAS Unigene10453 1.03 1.03 1.08 Scarecrow-like protein 15 WRKY CL6521.Contig1 2.84 1.66 2.35 Probable WRKY transcription factor 25 WRKY CL7324.Contig3 1.13 1.59 1.21 Probable WRKY transcription factor 35 WRKY CL791.Contig6 2.59 2.28 2.29 Probable WRKY transcription factor 40 bHLH Unigene12420 2.20 2.20 2.29 Phytochrome-interacting factor 3 MADS Unigene23315 2.17 3.01 2.27 MADS-box transcription factor 2.6 qRT-PCR验证基因表达差异
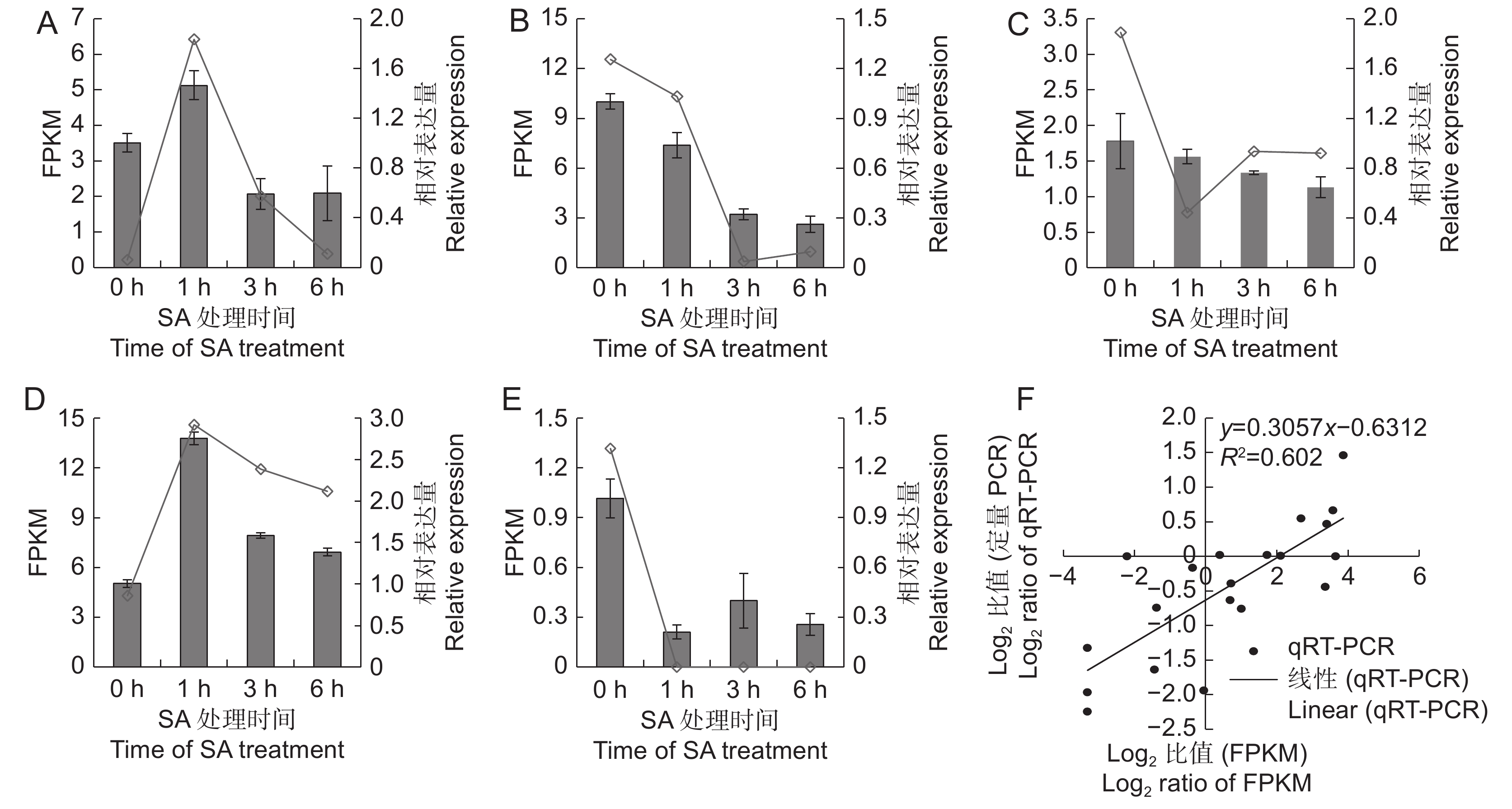
为了验证转录组测序对基因表达量分析的可靠性,随机选取CL7331.Contig2、Unigene1886、CL985.Contig1、CL5931.Contig2和CL379.Contig3等5个基因,利用qRT-PCR方法检测其在不同样本中的相对表达量。结果表明(图5),5个基因的定量结果与转录组获得的FPKM值变化趋势基本一致,其中CL7331.Contig2和CL5931.Contig2为SA处理后上调表达的基因,Unigene1886、CL985.Contig1和CL379.Contig3为SA处理后下调表达的基因。qRT-PCR与转录组测序的Pearson相关系数为0.602,相关性达显著水平,说明利用转录组测序分析SA处理后的基因表达量结果可靠。
![]() 图 5 差异表达基因的qRT-PCR验证A ~ E分别为CL7331.Contig2、Unigene1886、CL985.Contig1、CL5931.Contig2和CL379.Contig3的FPKM值与相对表达量;F:FPKM值与qRT-PCR的相关性分析。Figure 5. Validation of DEGs by qRT-PCRA–E: Represent expression and FPKM values of CL7331.Contig2, Unigene1886, CL985.Contig1, CL5931.Contig2, and CL379.Contig3; F: Correlation analysis between FPKM and qRT-PCR data.
图 5 差异表达基因的qRT-PCR验证A ~ E分别为CL7331.Contig2、Unigene1886、CL985.Contig1、CL5931.Contig2和CL379.Contig3的FPKM值与相对表达量;F:FPKM值与qRT-PCR的相关性分析。Figure 5. Validation of DEGs by qRT-PCRA–E: Represent expression and FPKM values of CL7331.Contig2, Unigene1886, CL985.Contig1, CL5931.Contig2, and CL379.Contig3; F: Correlation analysis between FPKM and qRT-PCR data.3. 讨论
植物次生代谢产物的积累既受自身遗传因素的控制,也受生长过程中生物与非生物环境条件的影响。一般而言,若药用植物的次生代谢产物在细胞中的含量相对较低,则会制约次生代谢产物的最终含量。近年来,人们常利用生物和非生物诱导子对植物进行处理,以提高植物特定次生代谢产物的生产[12]。如花生四烯酸(AA)、SA、茉莉酸甲酯(MeJA)和AgNO3均能够提高紫杉醇的含量[13]。50 µmol/L的乙烯利可以显著促进人参(Panax ginseng C. A. Meyer)根的生长和人参皂苷的积累[14]。诱导子提高苯乙醇苷含量的研究亦有报道,如外源添加Ag + 和腐胺均可以提高肉苁蓉(Cistanche deserticola Y. C. Ma)细胞培养物中松果菊苷和毛蕊花糖苷的含量[15]。本课题组前期研究发现,在地黄毛状根培养的培养基中添加25 μmol/L的SA可将毛蕊花糖苷的含量提高2.28倍[10]。本研究发现,叶面喷施100 μmol/L的SA可显著提高地黄叶片和块根中毛蕊花糖苷的含量,在块根中最高可提高1.4倍,说明在大田中叶面喷施诱导子可有效提高地黄块根中的次生代谢产物,有助于改善中药材的品质。
转录组测序分析不但可以高通量地获得基因表达的有关信息,还能够揭示基因表达与生命现象之间的内在联系,从而表征生命体的生理活动规律并确定其代谢特征[16]。目前,转录组测序不仅用于模式植物和大田作物生长发育及逆境胁迫响应关键基因的筛选[17-19],在药用植物次生代谢产物合成调控的结构基因和转录因子基因的挖掘中也有广泛应用[20-22]。由于地黄为同源四倍体物种,其基因组测序虽有报道[23],但作为参考基因组仍存在一些问题。因此,本研究利用课题组前期获得的地黄根、叶转录组为参考基因集进行分析,发现测序数据的特异匹配率偏低(50%左右),与地黄毛状根转录组测序的结果[10]类似,可能与其为同源四倍体物种有关。地黄叶片表面喷施SA后,上调表达的基因数少于下调表达,与SA处理的地黄毛状根结果[10]不同,这可能与本研究以大田地黄材料进行SA处理有关。本研究还发现,利用RNA-seq分析基因表达与qRT-PCR分析的结果相关系数仅为0.602,虽然达到显著相关,但未达到极显著相关水平,可能与地黄的基因组较大(约2.6 Gb),而采用RNA-seq测序获得的数据量较小有关。因此,对于基因组较大的物种,建议提高RNA-seq测序的深度,以获得更多的基因表达信息,提高基因表达量分析的准确性。
毛蕊花糖苷的生物合成途径目前已经比较清楚,其羟基酪醇基团来源于酪氨酸途径,咖啡酰基团来源于苯丙氨酸途径[24]。本课题组进一步推导、优化了毛蕊花糖苷的生物合成途径,认为毛蕊花糖苷是由羟基酪醇苷和咖啡酰辅酶A在莽草酸邻羟基肉桂酰转移酶(HCT)/毛蕊花糖苷合酶(AcS)和UGT的催化下合成[10]。周延清等[25] 基于地黄代谢组学分析获得了KEGG途径中的香豆酸-3-羟化酶(C3H),并克隆了其全长编码序列。李欣容等[26]根据SA处理下毛状根中催化酶基因的表达特性,鉴定并克隆了响应SA诱导的毛蕊花糖苷合酶基因RgAcS1。Yang等[27] 鉴定了4个酪氨酸脱羧酶(TyDC)基因,遗传转化发现过量表达RgTyDC2和RgTyDC4的地黄块根、纤维根、茎、嫩叶和成熟叶中的毛蕊花糖苷含量均显著高于野生型。Wang等[28]筛选了1个响应SA和H2O2诱导的WRKY转录因子基因RgWRKY37,功能研究发现RgWRKY37过量表达的毛状根转化体中毛蕊花糖苷和总苯乙醇苷的含量均显著高于对照。本研究发现,在SA处理的地黄块根中2个ALDH基因、1个UGT基因和2个PPO基因均上调表达,可能与块根中毛蕊花糖苷的含量增加有关。同时,在SA处理后的3个时间点,编码WKRY、NAC和AP2-EREBP等转录因子的20个基因均显著上调表达,其中RgWRKY37的表达量均明显增加。本研究结果为进一步探讨SA诱导毛蕊花糖苷合成的分子机理奠定了基础。
1 1)如需查阅附表内容请登录《植物科学学报》网站(http://www.plantscience.cn)查看本期文章。 -
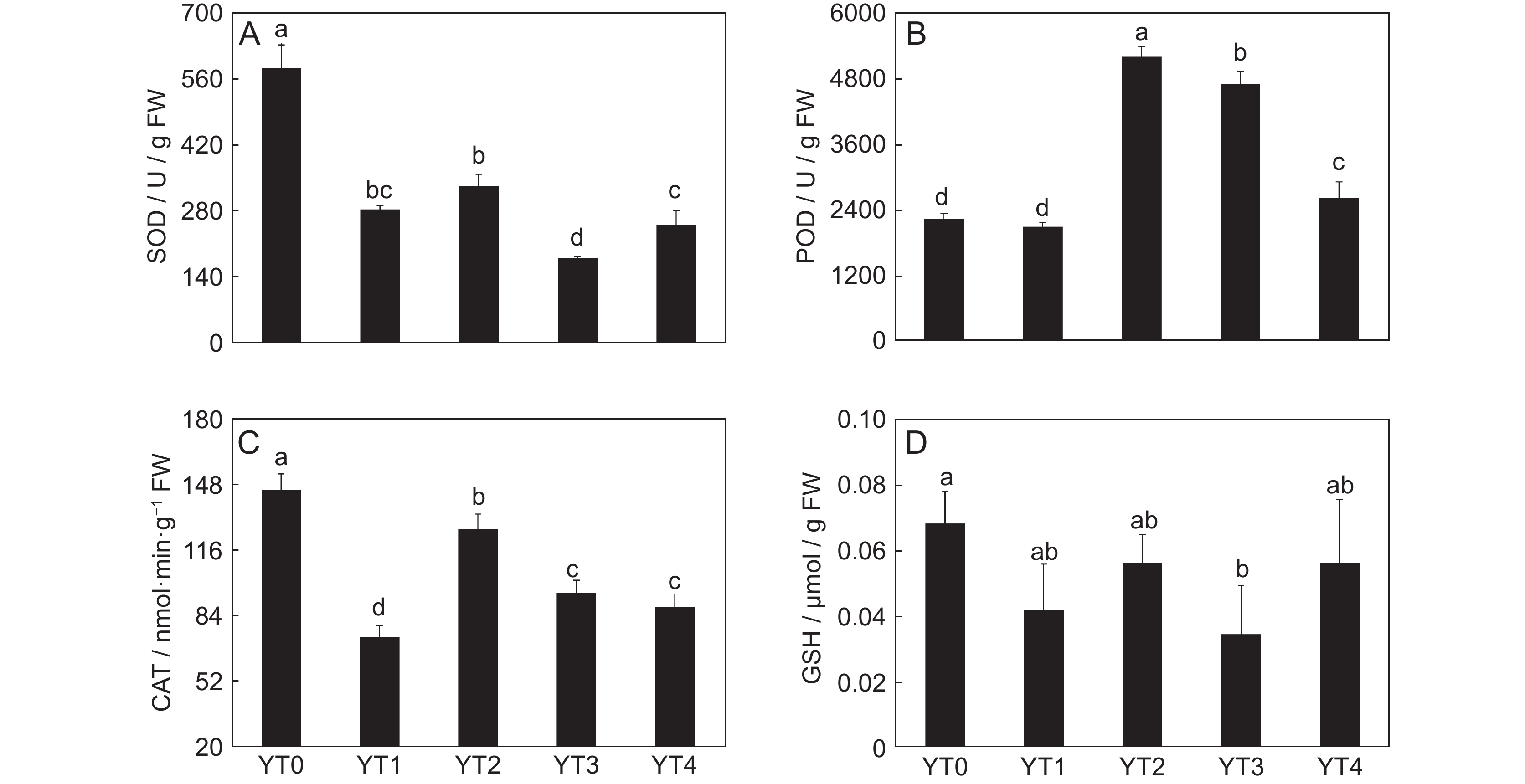
图 1 车前在Cd胁迫下超氧化物歧化酶SOD(A)、过氧化物酶POD(B)、过氧化氢酶CAT(C)活性和还原型谷胱甘肽GSH含量(D)的变化
YT0、YT1、YT2、YT3和YT4分别表示对照组及3、10、50、100 mg/kg Cd胁迫组。不同小写字母表示在0.05水平显著差异。
Figure 1. Changes in SOD (A), POD (B), CAT (C), and GSH (D) in Plantago asiatica under Cd stress
YT0, YT1, YT2, YT3, and YT4 represent CK, 3, 10, 50, and 100 mg/kg Cd stress groups, respectively. Different lowercase letters indicate significant differences at 0.05 level.
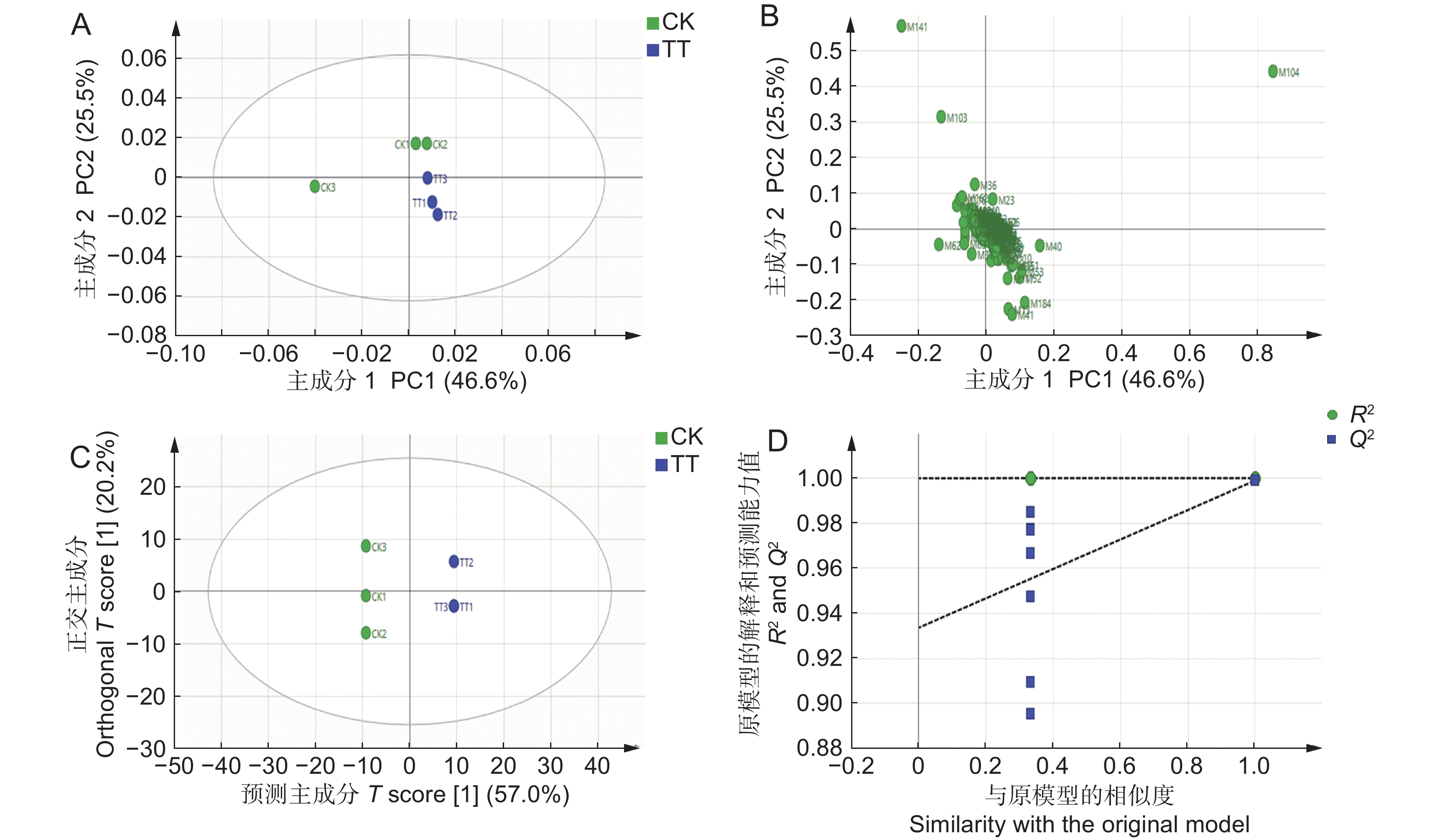
图 2 PCA得分散点图(A)、PCA载荷图(B)、OPLS-DA得分散点图(C)以及OPLS-DA模型的置换检验结果(D)
CK 和 TT 分别表示对照组和 50 mg/kg Cd 胁迫组。
Figure 2. Scatter plot of PCA scores (A), PCA load plot (B), scatter plot of OPLS-DA (C), and OPLS-DA model permutation test results (D)
CK and TT represent control and 50 mg/kg Cd stress groups, respectively.
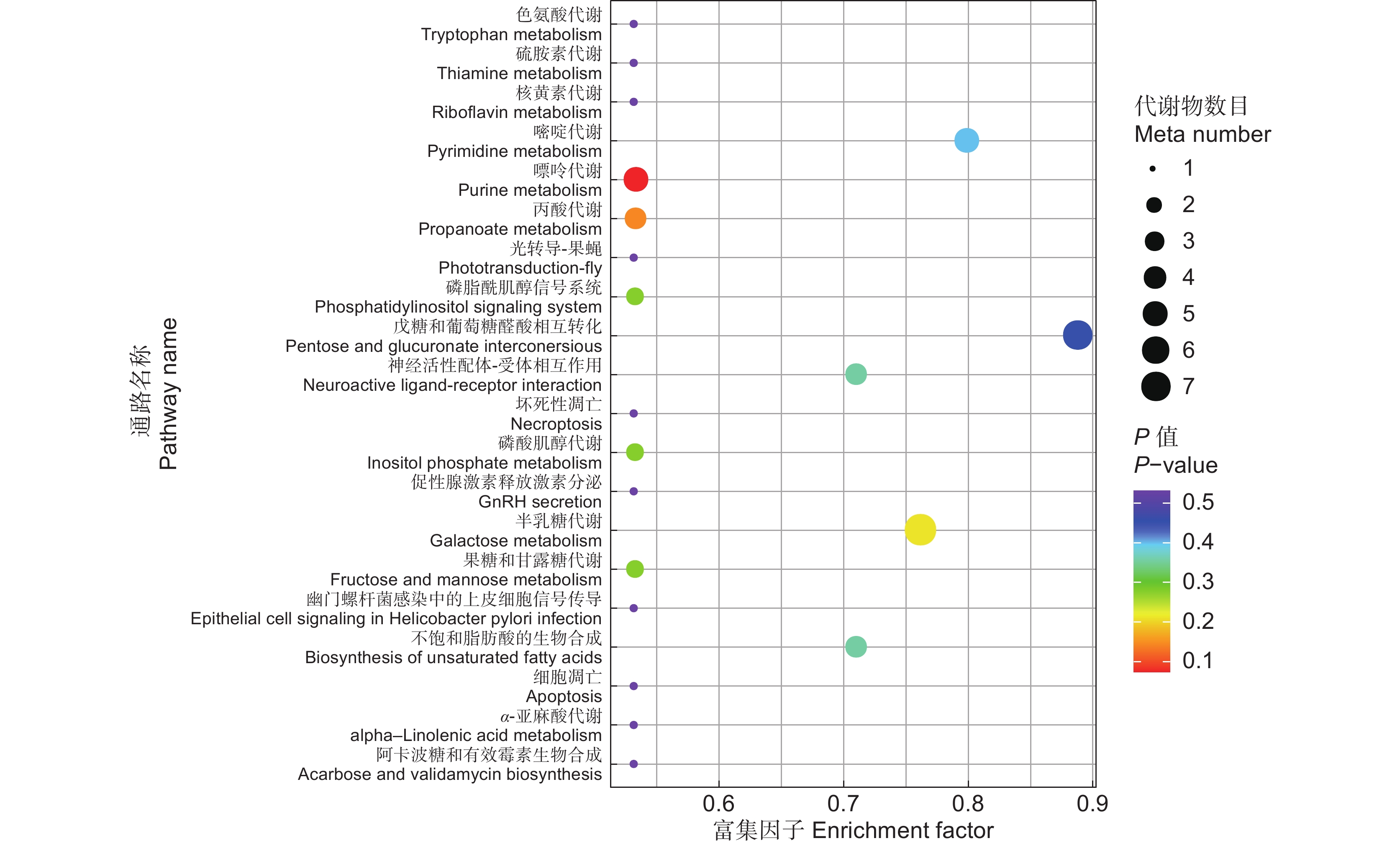
表 1 KEGG 通路富集详情
Table 1 KEGG pathway enrichment
KEGG 通路
KEGG pathway代谢物数目
Number of metabolites通路 ID
Pathway IDP 值
P-value化合物
Compound嘌呤代谢 4 map00230 0.07 cpd:C00037甘氨酸
cpd:C00242鸟嘌呤
cpd:C00064 L-谷氨酰胺
cpd:C00086尿素丙酸代谢 3 map00640 0.14 cpd:C05984 2-羟基丁酸
cpd:C05235 羟基丙酮
cpd:C00099 β-丙氨酸半乳糖代谢 7 map00052 0.21 cpd:C00984 α-D-半乳糖
cpd:C00103 D-葡萄糖1-磷酸
cpd:C00795 D-塔格糖
cpd:C00137肌醇
cpd:C05400蜜二糖
cpd:C05400蜜二糖
cpd:C00984 α-D-半乳糖磷酸肌醇代谢 2 map00562 0.28 cpd:C03546 D-肌醇4-磷酸
cpd:C00137肌醇磷脂酰肌醇信号系统 2 map04070 0.28 cpd:C03546 D-肌醇4-磷酸
cpd:C00137肌醇果糖和甘露糖代谢 2 map00051 0.28 cpd:C01487 D-阿洛糖
cpd:C00392甘露糖醇不饱和脂肪酸的生物合成 3 map01040 0.36 cpd:C00712油酸
cpd:C06427 α-亚麻酸
cpd:C01530硬脂酸神经活性配体-受体相互作用 3 map04080 0.36 cpd:C00037甘氨酸
cpd:C00099 β-丙氨酸
cpd:C00334 4-氨基丁酸嘧啶代谢 4 map00240 0.40 cpd:C00064 L-谷氨酰胺
cpd:C02067假尿苷
cpd:C00086尿素
cpd:C00099 β-丙氨酸戊糖和葡萄糖醛酸相互转化 6 map00040 0.45 cpd:C00103 D-葡萄糖-1-磷酸
cpd:C00514 D-甘露酸盐
cpd:C00502 D-木糖酸盐
cpd:C00800 L-古洛糖酸
cpd:C00474核糖醇
cpd:C00167 UDP-葡萄糖醛酸色氨酸代谢 1 map00380 0.53 cpd:C05659 5-甲氧基色胺 硫胺素代谢 1 map00730 0.53 cpd:C00037甘氨酸 核黄素代谢 1 map00740 0.53 cpd:C00474核糖醇 光转导-果蝇 1 map04745 0.53 cpd:C01530硬脂酸 坏死性凋亡 1 map04217 0.53 cpd:C00319鞘氨醇 促性腺激素释放激素分泌 1 map04929 0.53 cpd:C00334 4-氨基丁酸 幽门螺杆菌感染中的上皮细胞信号传导 1 map05120 0.53 cpd:C00086尿素 细胞凋亡 1 map04210 0.53 cpd:C00319鞘氨醇 α-亚麻酸代谢 1 map00592 0.53 cpd:C06427 α-亚麻酸 阿卡波糖和有效霉素生物合成 1 map00525 0.53 cpd:C00103 D-葡萄糖 1-磷酸 -
[1] 张雪芹,曲玮,梁敬钰. 车前草化学成分和药理作用研究进展[J]. 海峡药学,2013,25(11):1−8. Zhang XQ,Qu W,Liang JY. Research progresses on chemical constituents and pharmacological activities of Plantago spp.[J]. Strait Pharmaceutical Journal,2013,25 (11):1−8.
[2] 国家药典委员会. 中华人民共和国药典: 一部[M]. 北京: 中国医药科技出版社, 2020: 69−70. [3] Cui L,Wang XN,Li J,Gao XY,Zhang JW,Liu ZT. Ecological and health risk assessments and water quality criteria of heavy metals in the Haihe River[J]. Environ Pollut,2021,290:117971. doi: 10.1016/j.envpol.2021.117971
[4] Mahvi AH,Eslami F,Baghani AN,Khanjani N,Yaghmaeian K,Mansoorian HJ. Heavy metal pollution status in soil for different land activities by contamination indices and ecological risk assessment[J]. Int J Environ Sci Technol,2022,19 (8):7599−7616. doi: 10.1007/s13762-022-03960-z
[5] Bahloul M. Pollution characteristics and health risk assessment of heavy metals in dry atmospheric deposits from Sfax solar saltern area in southeast of Tunisia[J]. J Environ Health Sci Eng,2019,17 (2):1085−1105. doi: 10.1007/s40201-019-00423-5
[6] 环境保护部, 国土资源部. 全国土壤污染状况调查公报[EB/OL]. (2014-04-17) [2023-03-02]. http://www.gov.cn/foot/site1/20140417/782bcb88840814ba158d01.pdf. [7] 安婷婷,黄帝,王浩,张一,陈应龙. 植物响应镉胁迫的生理生化机制研究进展[J]. 植物学报,2021,56(3):347−362. doi: 10.11983/CBB20160 An TT,Huang D,Wang H,Zhang Y,Chen YL. Research advances in plant physiological and biochemical mechanisms in response to cadmium stress[J]. Chinese Bulletin of Botany,2021,56 (3):347−362. doi: 10.11983/CBB20160
[8] 文珂,刘文胜,赵运林. 重金属污染区与非污染区平车前生物量分配的比较[J]. 中南林业科技大学学报,2018,38(1):94−98. Wen K,Liu WS,Zhao YL. Comparisons of biomass allocation between Plantago depressa plants growing in heavy-metal polluted and nonpolluted soils[J]. Journal of Central South University of Forestry & Technology,2018,38 (1):94−98.
[9] Feng Z,Ji SY,Ping JF,Cui D. Recent advances in metabolomics for studying heavy metal stress in plants[J]. Trends Analyt Chem,2021,143:116402. doi: 10.1016/j.trac.2021.116402
[10] Ghatak A,Chaturvedi P,Weckwerth W. Metabolomics in plant stress physiology[J]. Adv Biochem Eng Biotechnol,2018,164:187−236.
[11] 刘荣鹏, 盛莎莎, 王晓云, 袁俊. 江西道地药用植物车前镉富集特点及其响应镉胁迫的转录组分析[J/OL]. 分子植物育种, 2023: 1−12. (2023-03-02). https://kns.cnki.net/kcms/detail//46.1068.s.20230228.1513.013.html. Liu RP, Sheng SS, Wang XY, Yuan J. Characteristics of cadmium enrichment of Jiangxi Daodi medicinal plant Plantago asiatica L. and its transcriptome analysis in response to cadmium stress[J/OL]. Molecular Plant Breeding, 2023: 1−12.(2023-03-02). https://kns.cnki.net/kcms/detail//46.1068.s.20230228.1513.013.html.
[12] Yuan J,Liu R,Sheng S,Fu H,Wang X. Integrated metabolomic and transcriptomic profiling revealed coping mechanisms of the edible and medicinal homologous plant Plantago asiatica L. cadmium resistance[J]. Open Life Sci,2022,17 (1):1347−1359. doi: 10.1515/biol-2022-0501
[13] 张凤,陈伟. 代谢组学在植物逆境生物学中的研究进展[J]. 生物技术通报,2021,37(8):1−11. Zhang F,Chen W. Research progress of metabolomics in plant stress biology[J]. Biotechnology Bulletin,2021,37 (8):1−11.
[14] 郭晋敏, 杨升, 陈秋夏, 刘星, 刘双双, 等. 极端低温胁迫下秋茄LC-MS代谢组学分析[J/OL]. 分子植物育种, 2021: 1−16. (2021-09-09). http://kns.cnki.net/kcms/detail/46.1068.S.20210909.1512.022.html. Guo JM, Yang S, Chen QX, Liu X, Liu SS, et al. Analysis of LC-MS metabolomics of Kandelia obovata under extreme low temperature stress[J/OL]. Molecular Plant Breeding, 2021: 1−16. (2021-09-09). http://kns.cnki.net/kcms/detail/46.1068.S.20210909.1512.022.html.
[15] 原静静,孙晓琛,栗锦鹏,杜弢,王惠珍. 基于LC-MS的干旱胁迫下党参代谢组学分析[J]. 中国实验方剂学杂志,2021,27(23):145−152. Yuan JJ,Sun XC,Li JP,Du T,Wang HZ. Metabolomics analysis of Codonopsis pilosula under drought stress based on LC-MS[J]. Chinese Journal of Experimental Traditional Medical Formulae,2021,27 (23):145−152.
[16] 朱建峰, 张会龙, 杨秀艳, 武海雯, 张华新. 盐胁迫下白榆种子萌发期代谢组学分析[J/OL]. 分子植物育种, 2021: 1-15. (2021-06-09). http://kns.cnki.net/kcms/detail/46.1068.S.20210609.1326.016.html. Zhu JF, Zhang HL, Yang XY, Wu HW, Zhang HX. Metabonomic analysis of seed germination period of Ulmus pumila under salt stress[J/OL]. Molecular Plant Breeding, 2021: 1−15. (2021-06-09). http://kns.cnki.net/kcms/detail/46.1068.S.20210609.1326.016.html.
[17] Hrydziuszko O,Viant MR. Missing values in mass spectrometry based metabolomics:an undervalued step in the data processing pipeline[J]. Metabolomics,2012,8 (S1):161−174. doi: 10.1007/s11306-011-0366-4
[18] Dunn WB,Broadhurst D,Begley P,Zelena E,Francis-Mcintyre S,et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry[J]. Nat Protoc,2011,6 (7):1060−1083. doi: 10.1038/nprot.2011.335
[19] Jolliffe IT,Cadima J. Principal component analysis:a review and recent developments[J]. Phil Trans Roy Soc A Math Phys Eng Sci,2016,374 (2065):20150202.
[20] Wiklund S,Johansson E,Sjöström L,Mellerowicz EJ,Edlund U,et al. Visualization of GC/TOF-MS-based metabolomics data for identification of biochemically interesting compounds using OPLS class models[J]. Anal Chem,2008,80 (1):115−122. doi: 10.1021/ac0713510
[21] Robotti E, Marengo E. Chemometric multivariate tools for candidate biomarker identification: LDA, PLS-DA, SIMCA, Ranking-PCA[M]//Marengo E, Robotti E, eds. 2-D PAGE Map Analysis: Methods and Protocols. New York: Humana, 2016: 237−267.
[22] Wishart DS,Feunang YD,Marcu A,Guo AC,Liang K,et al. HMDB 4.0:the human metabolome database for 2018[J]. Nucleic Acids Res,2018,46 (D1):D608−D617. doi: 10.1093/nar/gkx1089
[23] Hattori M,Tanaka N,Kanehisa M,Goto S. SIMCOMP/SUBCOMP:chemical structure search servers for network analyses[J]. Nucleic Acids Res,2010,38 (S2):W652−W656.
[24] Kanehisa M,Sato Y,Kawashima M,Furumichi M,Tanabe M. KEGG as a reference resource for gene and protein annotation[J]. Nucleic Acids Res,2016,44 (D1):D457−D462. doi: 10.1093/nar/gkv1070
[25] Xia JG,Sinelnikov IV,Han B,Wishart DS. MetaboAnalyst 3.0-making metabolomics more meaningful[J]. Nucleic Acids Res,2015,43 (W1):W251−W257. doi: 10.1093/nar/gkv380
[26] Saccenti E,Hoefsloot HCJ,Smilde AK,Westerhuis JA,Hendriks MMWB. Reflections on univariate and multivariate analysis of metabolomics data[J]. Metabolomics,2014,10 (3):361−374. doi: 10.1007/s11306-013-0598-6
[27] Das K,Roychoudhury A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants[J]. Front Environ Sci,2014,2:53.
[28] Zhou XR,Joshi S,Patil S,Khare T,Kumar V. Reactive oxygen,nitrogen,carbonyl and sulfur species and their roles in plant abiotic stress responses and tolerance[J]. J Plant Growth Regul,2022,41 (1):119−142. doi: 10.1007/s00344-020-10294-y
[29] 全芮萍,陈建福,张蕾,许明志,杨瑞芳,等. 抗氧化酶和植物螯合肽对苎麻重金属Cd胁迫的应答[J]. 热带作物学报,2022,43(5):1023−1031. doi: 10.3969/j.issn.1000-2561.2022.05.017 Quan RP,Chen JF,Zhang L,Xu MZ,Yang RF,et al. Responses of ramie to antioxidant enzymes and plant chelating peptides to Cd stress[J]. Chinese Journal of Tropical Crops,2022,43 (5):1023−1031. doi: 10.3969/j.issn.1000-2561.2022.05.017
[30] 张金钰. 糯玉米苗期镉胁迫下转录组和代谢组分析[D]. 广州: 仲恺农业工程学院, 2020: 78. [31] Wang JC,Chen XF,Chu SH,You YM,Chi YW,et al. Comparative cytology combined with transcriptomic and metabolomic analyses of Solanum nigrum L. in response to Cd toxicity[J]. J Hazard Mater,2022,423:127168. doi: 10.1016/j.jhazmat.2021.127168
[32] 曹莹,李建东,赵天宏,郭伟. 镉胁迫对玉米生理生化特性的影响[J]. 农业环境科学学报,2007,26(S1):8−11. Cao Y,Li JD,Zhao TH,Guo W. Effects of Cd stress on physiological and biochemical traits of maize[J]. Journal of Agro-Environment Science,2007,26 (S1):8−11.
[33] 朱润华,贺忠群,王海霞,白胜,阳圣莹,蒋浩宏. 镉胁迫处理对水培苦苣幼苗生理响应及叶片超微结构的影响[J]. 西南农业学报,2021,34(6):1302−1308. Zhu RH,He ZQ,Wang HX,Bai S,Yang SY,Jiang HH. Effects of cadmium stress on physiological response and leaf ultrastructure of hydroponic Cichorium endivia L. seedling[J]. Southwest China Journal of Agricultural Sciences,2021,34 (6):1302−1308.
[34] 黄东华,麦淑华,仇曙,陈大清. 镉对堇叶碎米荠生长生理特性的影响[J]. 湖北农业科学,2022,61(5):87−90. Huang DH,Mai SH,Qiu S,Chen DQ. Effects of cadmium on growth and physiological characteristics of Cardamine violifolia[J]. Hubei Agricultural Sciences,2022,61 (5):87−90.
[35] 卢倩云,曹宇棽,陈友明,晏琼. 镉胁迫下油菜毛状根的生理响应及铁钾含量[J]. 应用与环境生物学报,2018,24(6):1382−1389. Lu QY,Cao YS,Chen YM,Yan Q. The physiological response and iron and potassium contents in the hairy roots of Brassica rape L. under cadmium stress[J]. Chinese Journal of Applied and Environmental Biology,2018,24 (6):1382−1389.
[36] Zhang QW,Song XM,Bartels D. Enzymes and metabolites in carbohydrate metabolism of desiccation tolerant plant[J]. Proteomes,2016,4 (4):40. doi: 10.3390/proteomes4040040
[37] 胡雪萍. 水稻响应镉胁迫代谢组学研究[D]. 南昌: 南昌大学, 2019: 78. [38] 汪怡文. 转录组和代谢组整合分析冬小麦镉胁迫响应的关键代谢通路[D]. 武汉: 华中农业大学, 2021: 81. [39] 谭飘飘. 芥菜对镉胁迫的生理代谢响应及外源脯氨酸的调控作用研究[D]. 长沙: 中南林业科技大学, 2021: 105. [40] 胡立群,徐庆国. 植物非生物胁迫代谢组学研究进展[J]. 作物研究,2014,28(4):428−434. doi: 10.3969/j.issn.1001-5280.2014.04.24 Hu LQ,Xu QG. Review of current progress in the metabolomics for plant response to abiotic stress[J]. Crop Research,2014,28 (4):428−434. doi: 10.3969/j.issn.1001-5280.2014.04.24
[41] Yang QQ,Zhao DS,Liu QQ. Connections between amino acid metabolisms in plants:lysine as an example[J]. Front Plant Sci,2020,11:928. doi: 10.3389/fpls.2020.00928
[42] Hildebrandt TM. Synthesis versus degradation:directions of amino acid metabolism during Arabidopsis abiotic stress response[J]. Plant Mol Biol,2018,98 (1-2):121−135. doi: 10.1007/s11103-018-0767-0
[43] Rizhsky L,Liang HJ,Shuman J,Shulaev V,Davletova S,Mittler R. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress[J]. Plant Physiol,2004,134 (4):1683−1696. doi: 10.1104/pp.103.033431
[44] 袁俊,盛莎莎,刘荣鹏,王晓云. 镉胁迫对丹参生理特性和代谢特征的影响[J]. 植物科学学报,2022,40(3):408−417. doi: 10.11913/PSJ.2095-0837.2022.30408 Yuan J,Sheng SS,Liu RP,Wang XY. Effects of cadmium on Physiological characteristics and metabolic profiles of Salvia miltiorrhiza Bunge[J]. Plant Science Journal,2022,40 (3):408−417. doi: 10.11913/PSJ.2095-0837.2022.30408
[45] Yuan J,Liu R,Sheng S,Fu H,Wang X. Untargeted LC-MS/MS-Based metabolomic profiling for the edible and medicinal plant Salvia miltiorrhiza under different levels of cadmium stress[J]. Front Plant Sci,2022,13:889370. doi: 10.3389/fpls.2022.889370
[46] Kaplan F,Kopka J,Haskell DW,Zhao W,Schiller KC,et al. Exploring the temperature-stress metabolome of Arabidopsis[J]. Plant Physiol,2004,136 (4):4159−4168. doi: 10.1104/pp.104.052142
[47] Sun XM,Zhang JX,Zhang HJ,Ni YW,Zhang Q,et al. The responses of Arabidopsis thaliana to cadmium exposure explored via metabolite profiling[J]. Chemosphere,2010,78 (7):840−845. doi: 10.1016/j.chemosphere.2009.11.045
[48] Broeckling CD,Huhman DV,Farag MA,Smith JT,May GD,et al. Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism[J]. J Exp Bot,2005,56 (410):323−336. doi: 10.1093/jxb/eri058
[49] Parthasarathy A,Savka MA,Hudson AO. The synthesis and role of β-Alanine in plants[J]. Front Plant Sci,2019,10:921. doi: 10.3389/fpls.2019.00921
[50] Katahira R,Ashihara H. Dual function of pyrimidine metabolism in potato (Solanum tuberosum) plants:pyrimidine salvage and supply of β‐alanine to pantothenic acid synthesis[J]. Physiol Plant,2006,127 (1):38−43. doi: 10.1111/j.1399-3054.2006.00658.x
[51] 王佳钰,李萌,宋晓卉,李琦,齐秀芬,王兰兰. 代谢组学在植物重金属胁迫研究中的应用[J]. 生物化工,2020,6(2):128−132. doi: 10.3969/j.issn.2096-0387.2020.02.037 Wang JY,Li M,Song XH,Li Q,Qi XF,Wang LL. The application of metabonomics in study of plant under heavy metal stress[J]. Biological Chemical Engineering,2020,6 (2):128−132. doi: 10.3969/j.issn.2096-0387.2020.02.037
[52] Kim HU. Lipid metabolism in plants[J]. Plants,2020,9 (7):871. doi: 10.3390/plants9070871
[53] Zhao XC,Wei YL,Zhang JJ,Yang LY,Liu XY,et al. Membrane lipids’ metabolism and transcriptional regulation in maize roots under cold stress[J]. Front Plant Sci,2021,12:639132. doi: 10.3389/fpls.2021.639132
[54] 贾斌, 高龙飞, 张卫华, 勾峰, 李天杰, 等. 西瓜苗期干旱胁迫下的代谢组学分析[J/OL]. 分子植物育种, 2021: 1−15. (2021-11-20). http://kns.cnki.net/kcms/detail/46.1068.S.20211118.2148.007.html. Jia B, Gao LF, Zhang WH, Gou F, Li TJ, et al. Metabonomics analysis of watermelon seedlings under drought stress[J/OL]. Molecular Plant Breeding, 2021: 1−15. (2021-11-20). http://kns.cnki.net/kcms/detail/46.1068.S.20211118.2148.007.html.
[55] 方彦,曾秀存,马骊,孙柏林,武军艳,等. 低温胁迫下冬油菜陇油7号根部代谢组学分析[J]. 干旱地区农业研究,2021,39(4):80−85. doi: 10.7606/j.issn.1000-7601.2021.04.10 Fang Y,Zeng XC,Ma L,Sun BL,Wu JY,et al. Metabolomics profiling of Brassica campestris Longyou 7 roots under cold stress[J]. Agricultural Research in the Arid Areas,2021,39 (4):80−85. doi: 10.7606/j.issn.1000-7601.2021.04.10
[56] Sun L,Cao X,Tan C,Deng Y,Cai R,Peng X,et al. Analysis of the effect of cadmium stress on root exudates of Sedum plumbizincicola based on metabolomics[J]. Ecotox Environ Safe,2020,205:111152. doi: 10.1016/j.ecoenv.2020.111152
-
期刊类型引用(0)
其他类型引用(1)
-
其他相关附件
-
DOCX格式
盛莎莎附表1 点击下载(29KB)
-



 下载:
下载: