Characterization and phylogenetic analysis of the complete chloroplast genome of Dictamnus dasycarpus Turcz.
-
摘要:
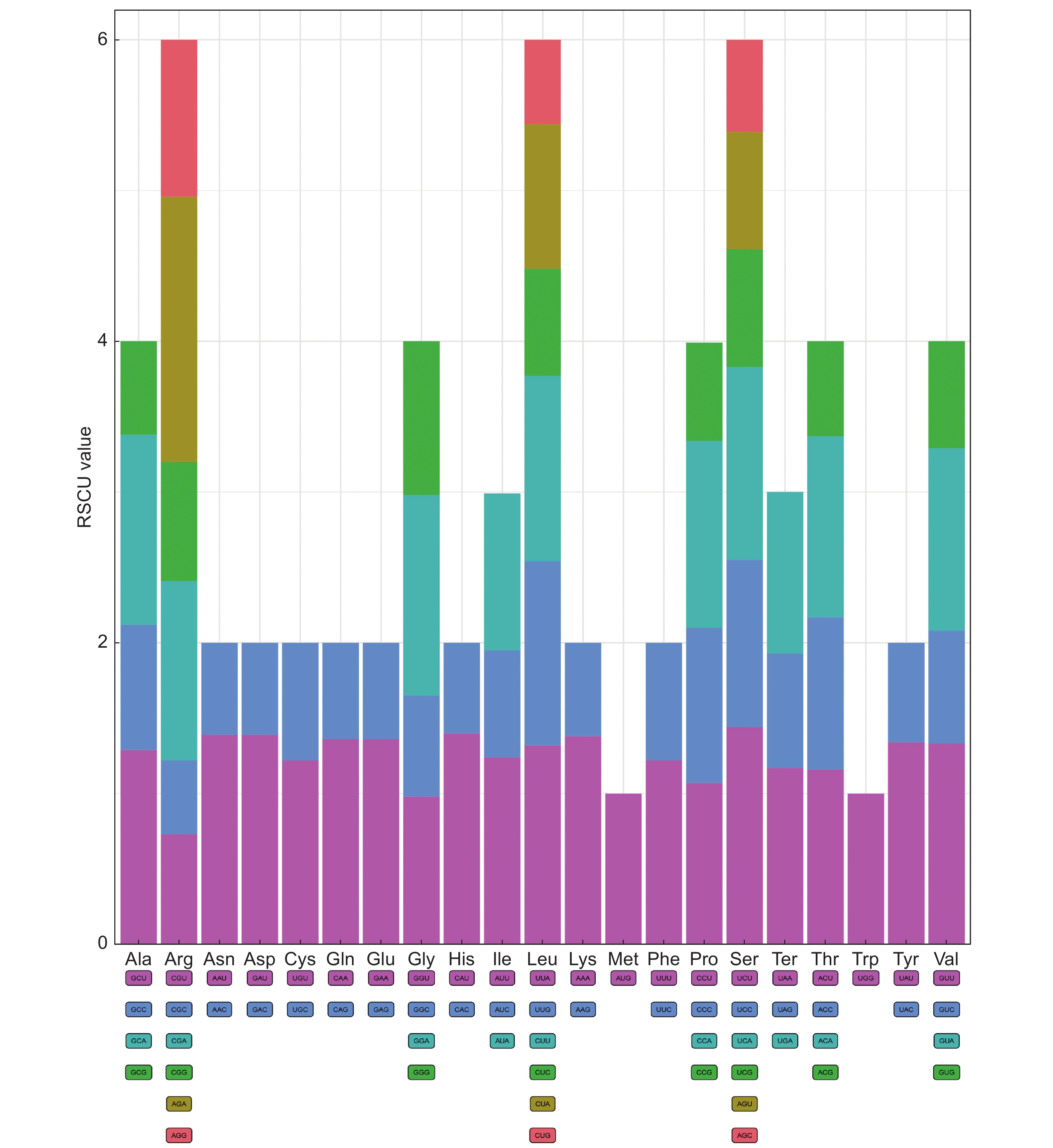
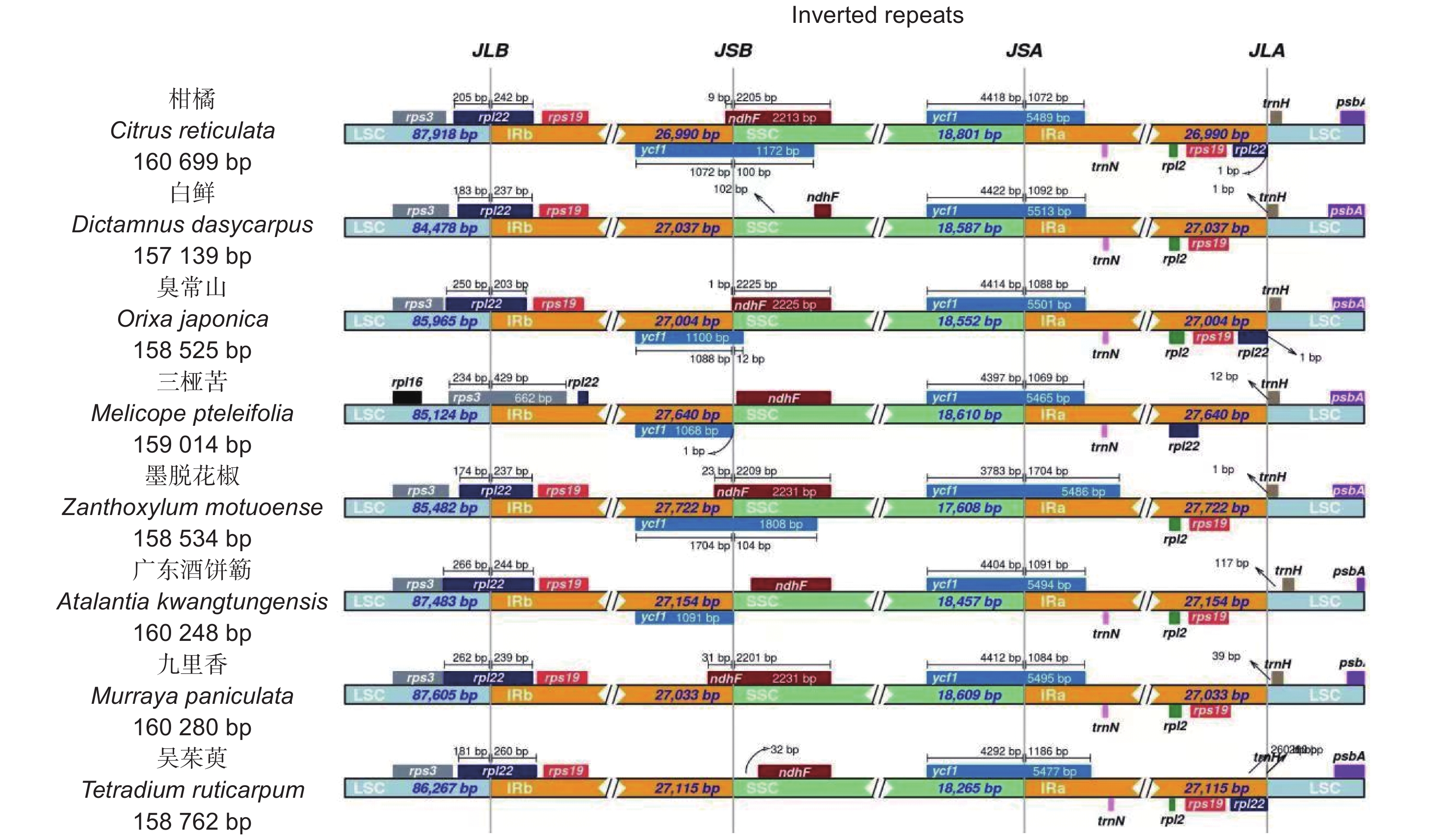
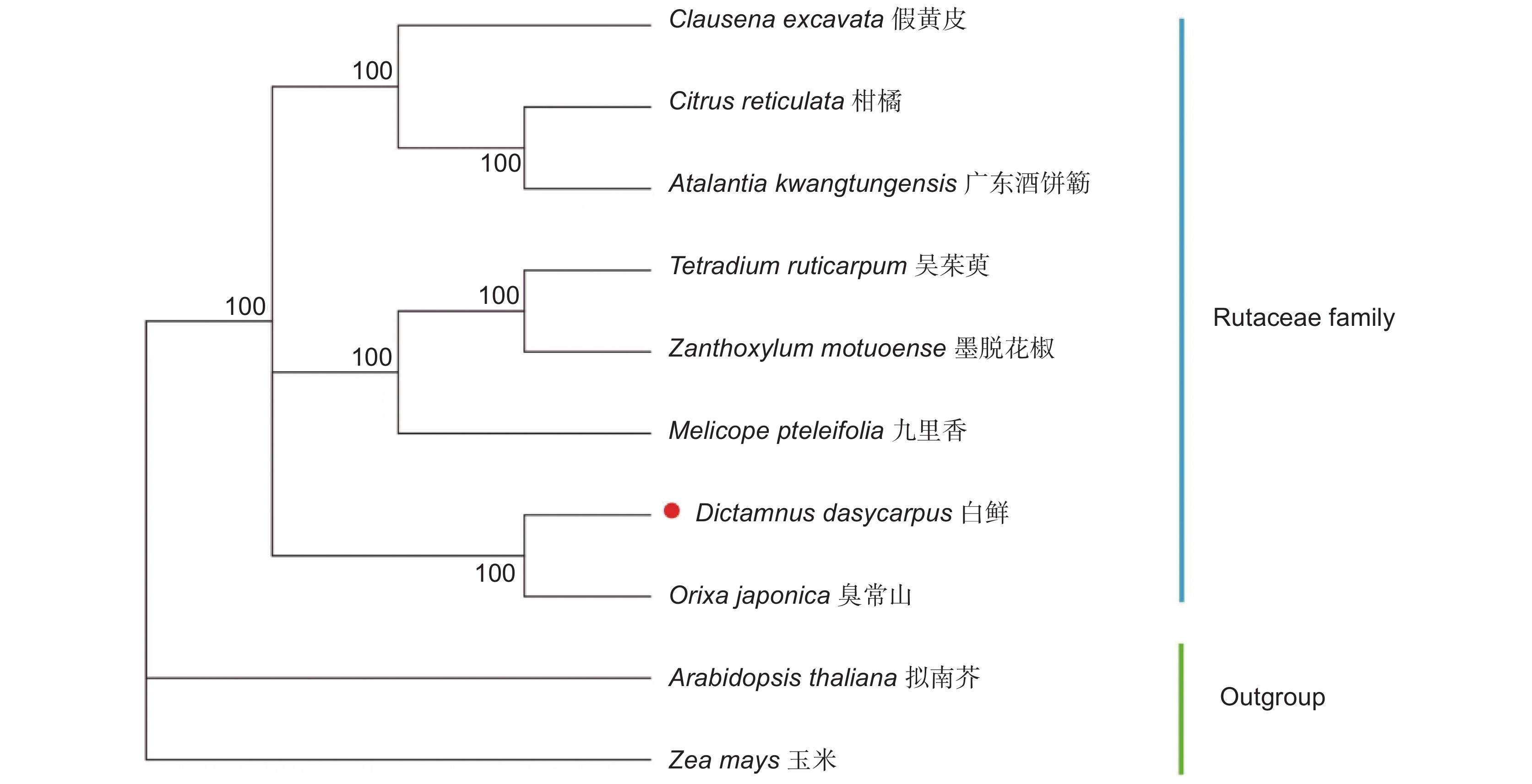
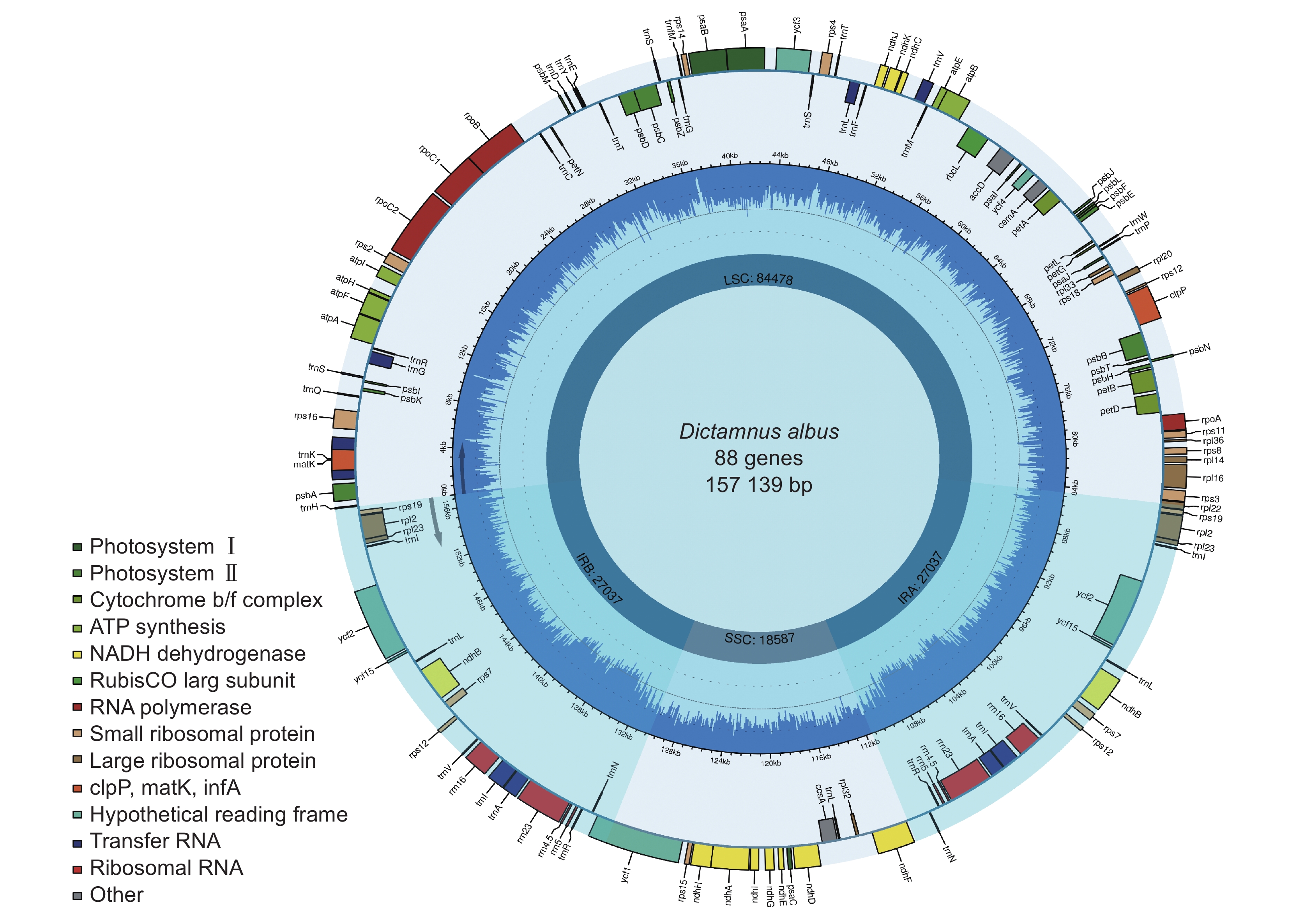
通过高通量测序技术获取白鲜(Dictamnus dasycarpus Turcz.)叶绿体全基因组序列,并采用生物信息学方法对其结构特征和系统发育关系进行了分析。结果显示,白鲜叶绿体基因组具有典型的环状四分体结构,总长度为157 139 bp,大单拷贝区和小单拷贝区长度分别为84 478 bp和18 587 bp,一对反向重复序列长度为27 037 bp,总GC含量为38.5%。共注释到132个基因,包括87个蛋白编码基因(PCGs)、8个rRNA和37个tRNA基因。计算相对同义密码子使用频率(RSCU),发现编码最多的密码子是亮氨酸(Leu/L),编码最少的密码子是半胱氨酸(Cys/C),RSCU值大于1的密码子有32个。白鲜叶绿体基因组共包含61个简单重复序列。通过对芸香科8种植物的叶绿体基因组结构进行比较,发现白鲜属与其他属的叶绿体基因组结构基本一致,并未发生重排和倒位现象。系统进化树分析结果表明,白鲜与臭常山(Orixa japonica Thunb.)的亲缘关系最近。
Abstract:The whole genome sequence of Dictamnus dasycarpus Turcz. chloroplasts was obtained by high-throughput sequencing, and its structural features and phylogenetic relationships were analyzed by bioinformatics. Results showed that the chloroplast genome had a typical circular tetrameric structure with a total length of 157 139 bp, a large and small single-copy region of 84 478 bp and 18 587 bp, respectively, a pair of inverted repeat sequences of 27 037 bp, and a total GC content of 38.5%. In total, 132 genes were annotated, including 87 protein-coding genes (PCGs), eight ribosomal RNA (rRNA) genes, and 37 transfer RNA (tRNA) genes. The frequency of relative synonymous codon use (RSCU) was calculated, showing that the most encoded codon was leucine (Leu/L) and the least encoded codon was cysteine (Cys/C), with 32 codons with RSCU values greater than 1. The D. dasycarpus chloroplast genome contained a total of 61 simple repetitive sequences. Compared with eight species of Rutaceae, the chloroplast genome structure was generally consistent between Dictamnus and other genera, with no rearrangements or inversions. Phylogenetic analysis showed that D. dasycarpus was most closely related to Orixa japonica Thunb.
-
Keywords:
- Dictamnus dasycarpus /
- Chloroplast genome /
- Rutaceae /
- Codon /
- Phylogeny
-
-
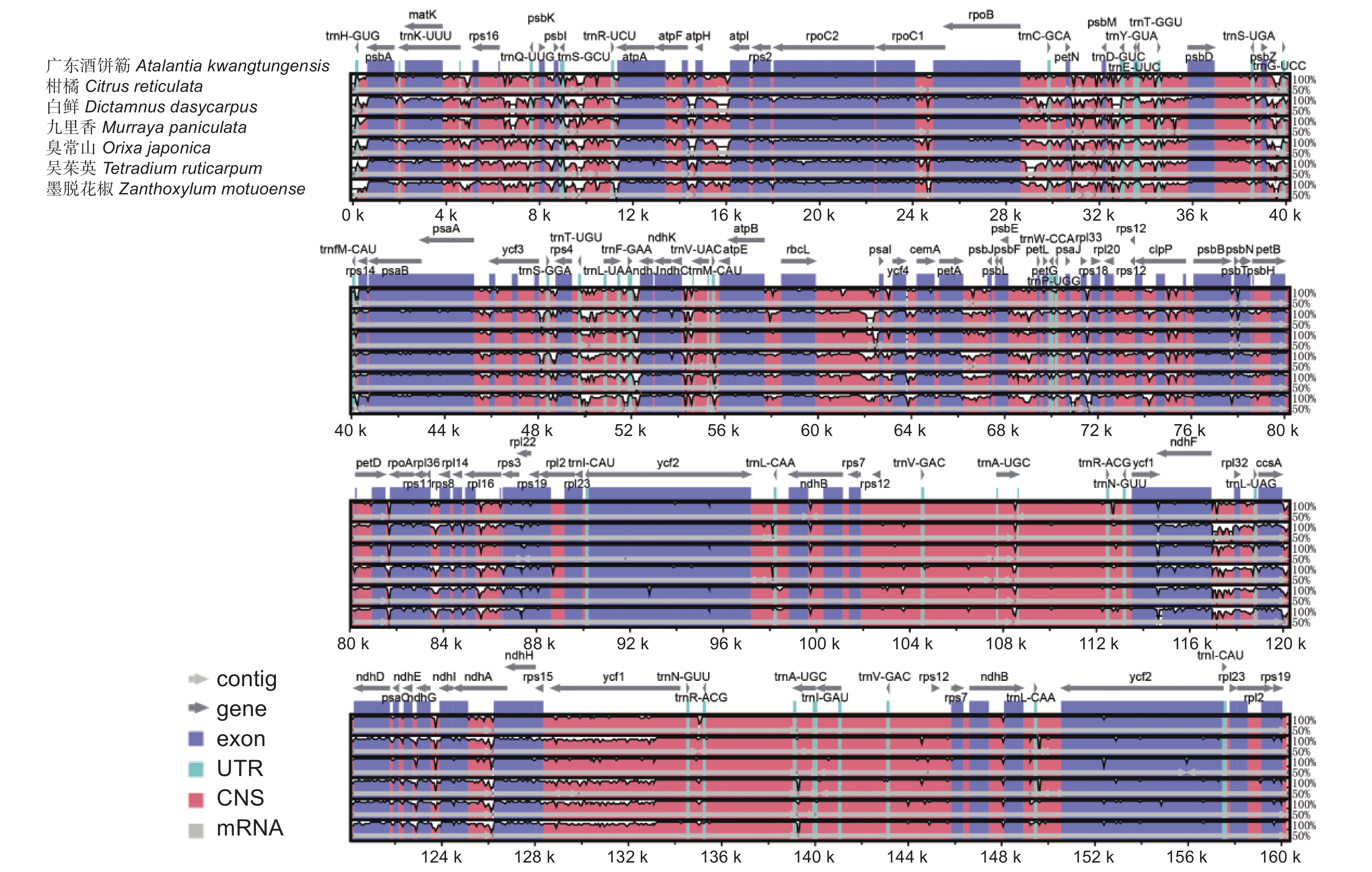
图 4 芸香科7种植物叶绿体基因组的可视化比对
使用广东酒饼簕为参考基因组。横轴表示比对的7个芸香科物种的叶绿体基因组。垂直比例表示百分比同一性,范围为 50% ~ 100%。不同颜色表示不同基因的功能。
Figure 4. Visual alignment of chloroplast genomes of seven species of Rutaceae
Atalantia kwangtungensis was used as a reference genome. Horizontal axis indicates seven aligned chloroplast genomes. Vertical scale represents percentage identity, ranging from 50%–100%. Different colors indicate functions of different genes.
表 1 白鲜叶绿体基因组碱基组成
Table 1 Base composition of chloroplast genome in Dictamnus dasycarpus
区域
RegionA / % T / % G / % C / % 长度
Length / bpGC / % 大单拷贝区(LSC) 30.9 32.2 17.9 19 84478 36.9 小单拷贝区(SSC) 33.7 33.2 16 17.1 18587 33.1 反向重复序列A(IRA) 28.7 28.4 22.2 20.7 27037 42.9 反向重复序列B(IRB) 28.4 28.7 20.7 22.2 27037 42.9 合计 30.4 31.1 18.9 19.6 157139 38.5 -
[1] 马孟莉,张薇,孟衡玲,王田涛,李春燕,卢丙越. 豆蔻属药用植物叶绿体基因组密码子偏性分析[J]. 中草药,2021,52(12):3661−3670. doi: 10.7501/j.issn.0253-2670.2021.12.024 Ma ML,Zhang W,Meng HL,Wang TT,Li CY,Lu BY. Codon bias analysis of chloroplast genome in medicinal plants of Amomum Roxb[J]. Chinese Traditional and Herbal Drugs,2021,52 (12):3661−3670. doi: 10.7501/j.issn.0253-2670.2021.12.024
[2] Ris H,Plaut W. Ultrastructure of DNA-containing areas in the chloroplast of Chlamydomonas[J]. J Cell Biol,1962,13 (3):383−391. doi: 10.1083/jcb.13.3.383
[3] Jansen RK,Raubeson LA,Boore JL,de Pamphilis CW,Chumley TW,et al. Methods for obtaining and analyzing whole chloroplast genome sequences[J]. Methods Enzymol,2005,395:348−384.
[4] 樊守金,郭秀秀. 植物叶绿体基因组研究及应用进展[J]. 山东师范大学学报(自然科学版),2022,37(1):22−31. doi: 10.3969/j.issn.1001-4748.2022.01.003 Fan SJ,Guo XX. Advances in research and application of plant chloroplast genome[J]. Journal of Shandong Normal University (Natural Science)
,2022,37 (1):22−31. doi: 10.3969/j.issn.1001-4748.2022.01.003 [5] 刘雷,郭丽娜,于春磊,孙宇,刘吉成. 白鲜皮化学成分及药理活性研究进展[J]. 中成药,2016,38(12):2657−2665. [6] 王英华. 白鲜在北方寒地的栽培与应用研究[J]. 农业科技与装备,2021(1):7−8. [7] 贾晓龙,刘卉,李莹,南桂仙. 白鲜生物学特性、化学组分与药理作用的研究现状[J]. 黑龙江科学,2018,9(20):42−43. [8] 李然红,王立凤,纪春艳,柴军红. 白鲜药用价值研究进展[J]. 安徽农业科学,2013,41(1):81−82. doi: 10.3969/j.issn.0517-6611.2013.01.034 [9] 阮淑明. 芸香科5种保健植物花粉形态扫描电镜观察[J]. 电子显微学报,2021,40(6):747−752. doi: 10.3969/j.issn.1000-6281.2021.06.016 Ruan SM. SEM observation on pollen morphology of five health plants in Rutaceae[J]. Journal of Chinese Electron Microscopy Society,2021,40 (6):747−752. doi: 10.3969/j.issn.1000-6281.2021.06.016
[10] 刘文哲,胡正海. 中国芸香科植物叶分泌囊比较解剖学研究[J]. 植物分类学报,1998,36(2):119−127. Liu WZ,Hu ZH. Comparative anatomy of secretory cavities in leaves of the Rutaceae in China[J]. Journal of Systematics and Evolution,1998,36 (2):119−127.
[11] 王闽予,邓淙友,陈康梅,罗文安,陈万发,等. 6种芸香科药材的特征图谱及含量对比研究[J]. 广东药科大学学报,2021,37(5):61−71. Wang MY,Deng CY,Chen KM,Luo WA,Chen WF,et al. Study on the characteristic chromatograms and the content of six kinds of Rutaceae herbs[J]. Journal of Guangdong Pharmaceutical University,2021,37 (5):61−71.
[12] Attitalla IH. Modified CTAB method for high quality genomic DNA extraction from medicinal plants[J]. Pakistan Journal of Biological Sciences,2011,14 (21):998−999. doi: 10.3923/pjbs.2011.998.999
[13] Jin JJ,Yu WB,Yang JB,Song Y,de Pamphilis CW,et al. GetOrganelle:a fast and versatile toolkit for accurate de novo assembly of organelle genomes[J]. Genome Biol,2020,21 (1):241. doi: 10.1186/s13059-020-02154-5
[14] Al-Qurainy F,Khan S,Tarroum M,Al-Hemaid FM,Ali MA. Molecular authentication of the medicinal herb Ruta graveolens (Rutaceae) and an adulterant using nuclear and chloroplast DNA markers[J]. Genet Mol Res,2011,10 (4):2806−2816. doi: 10.4238/2011.November.10.3
[15] Tillich M,Lehwark P,Pellizzer T,Ulbricht-Jones ES,Fischer A,et al. GeSeq-versatile and accurate annotation of organelle genomes[J]. Nucleic Acids Res,2017,45 (W1):W6−W11. doi: 10.1093/nar/gkx391
[16] Wickham H. ggplot2: Elegant Graphics for Data Analysis[M]. New York: Springer-Verlag, 2016: 1−10.
[17] Frazer KA,Pachter L,Poliakov A,Rubin EM,Dubchak I. VISTA:computational tools for comparative genomics[J]. Nucleic Acids Res,2004,32 (S2):W273−W279.
[18] Rozewicki J,Li SL,Amada KM,Standley DM,Katoh K. MAFFT-DASH:integrated protein sequence and structural alignment[J]. Nucleic Acids Res,2019,47 (W1):W5−W10.
[19] Kumar S,Stecher G,Li M,Knyaz C,Tamura K. MEGA X:molecular evolutionary genetics analysis across computing platforms[J]. Mol Biol Evol,2018,35 (6):1547−1549. doi: 10.1093/molbev/msy096
[20] Ling LZ,Zhang SD. The complete chloroplast genome of the multipurpose and traditional herb,Ruta graveolens L.[J]. Mitochondrial DNA Part B Resour,2019,4 (2):3661−3662. doi: 10.1080/23802359.2019.1678426
[21] Shinozaki K,Ohme M,Tanaka M,Wakasugi T,Hayashida N,et al. The complete nucleotide sequence of the tobacco chloroplast genome:its gene organization and expression[J]. EMBO J,1986,5 (9):2043−2049. doi: 10.1002/j.1460-2075.1986.tb04464.x
[22] 王雪芹, 宋卫武, 高晓燕, 林悦, 郭慧. 芸香科植物叶绿体基因组结构和系统发育分析[J/OL]. 分子植物育种. (2021-04-19). http://kns.cnki.net/kcms/detail/46.1068.S.20210419.1547.019.html. [23] Wang WB,Yu H,Wang JH,Lei WJ,Gao JH,et al. The complete chloroplast genome sequences of the medicinal plant forsythia suspensa (Oleaceae)[J]. Int J Mol Sci,2017,18 (11):2288. doi: 10.3390/ijms18112288
[24] Su HJ,Hogenhout SA,Al-Sadi AM,Kuo CH. Complete chloroplast genome sequence of Omani lime (Citrus aurantiifolia) and comparative analysis within the rosids[J]. PLoS One,2014,9 (11):e113049. doi: 10.1371/journal.pone.0113049
[25] Zhu ZX,Wang JH,Sun XZ,Zhao KK,Wang HF. Complete plastome sequence of Atalantia kwangtungensis (Rutaceae):an endemic "near threatened" shrub in South China[J]. Mitochondrial DNA Part B Resour,2018,3 (2):730−731. doi: 10.1080/23802359.2018.1483764
[26] Appelhans MS,Krohm S,Manafzadeh S,Wen J. Phylogenetic placement of Psilopeganum,a rare monotypic genus of Rutaceae (the citrus family) endemic to China[J]. J Syst Evol,2016,54 (5):535−544. doi: 10.1111/jse.12208
[27] Ying ZQ,Yan MX,Zhou MJ,He XY,Cheng RB. Characterization of the complete chloroplast genome of the medicinal plant Orixa japonica (Rutaceae) in Zhejiang Province and its phylogenetic analysis within family Rutaceae[J]. Mitochondrial DNA Part B Resour,2021,6 (6):1734−1736. doi: 10.1080/23802359.2021.1931511
[28] Muellner-Riehl AN,Weeks A,Clayton JW,Buerki S,Nauheimer L,et al. Molecular phylogenetics and molecular clock dating of Sapindales based on plastid rbcL,atpB and trnL-trnF DNA sequences[J]. Taxon,2016,65 (5):1019−1036. doi: 10.12705/655.5
[29] Grant M,Blackmore S,Morton C. Pollen morphology of the subfamily Aurantioideae (Rutaceae)[J]. Grana,2000,39 (1):8−20. doi: 10.1080/00173130150503768
[30] 贾富霞,王秀娟,罗容. 酸橙枳实HPLC指纹图谱方法研究及6种黄酮类药效组分的含量测定[J]. 中华中医药学刊,2016,34(12):2917−2920. Jia FX,Wang XJ,Luo R. Study on HPLC chromatographic fingerprint method for simultaneous determination of six flavonoid components in aurantii fructus immaturus from cirtus aurantium L.[J]. Chinese Archives of Traditional Chinese Medicine,2016,34 (12):2917−2920.
[31] Appelhans MS,Reichelt N,Groppo M,Paetzold C,Wen J. Phylogeny and biogeography of the pantropical genus Zanthoxylum and its closest relatives in the proto-Rutaceae group (Rutaceae)[J]. Mol Phylogenet Evol,2018,126:31−44. doi: 10.1016/j.ympev.2018.04.013
[32] 桂茜,蒋昊,阮颖. 7种芸香科植物的RAPD分析[J]. 安徽农业科学,2016,44(30):101−103. Gui X,Jiang H,Ruan Y. RAPD analysis of seven species of rutaceae plants[J]. Journal of Anhui Agricultural Sciences,2016,44 (30):101−103.
[33] 徐世荣,陈燕琼,潘东明,潘鹤立. ‘六月早’蜜柚叶绿体基因组及其特征分析[J]. 热带作物学报,2021,42(5):1223−1230. doi: 10.3969/j.issn.1000-2561.2021.05.004 Xu SR,Chen YQ,Pan DM,Pan HL. Chloroplast genome sequence and characteristics analysis of Citrus maxima ‘Liuyuezao’[J]. Chinese Journal of Tropical Crops,2021,42 (5):1223−1230. doi: 10.3969/j.issn.1000-2561.2021.05.004



 下载:
下载: