Genome-wide identification and characterization of MADS-box gene family of Rhododendron griersonianum Balf. f. et Forrest
-
摘要:
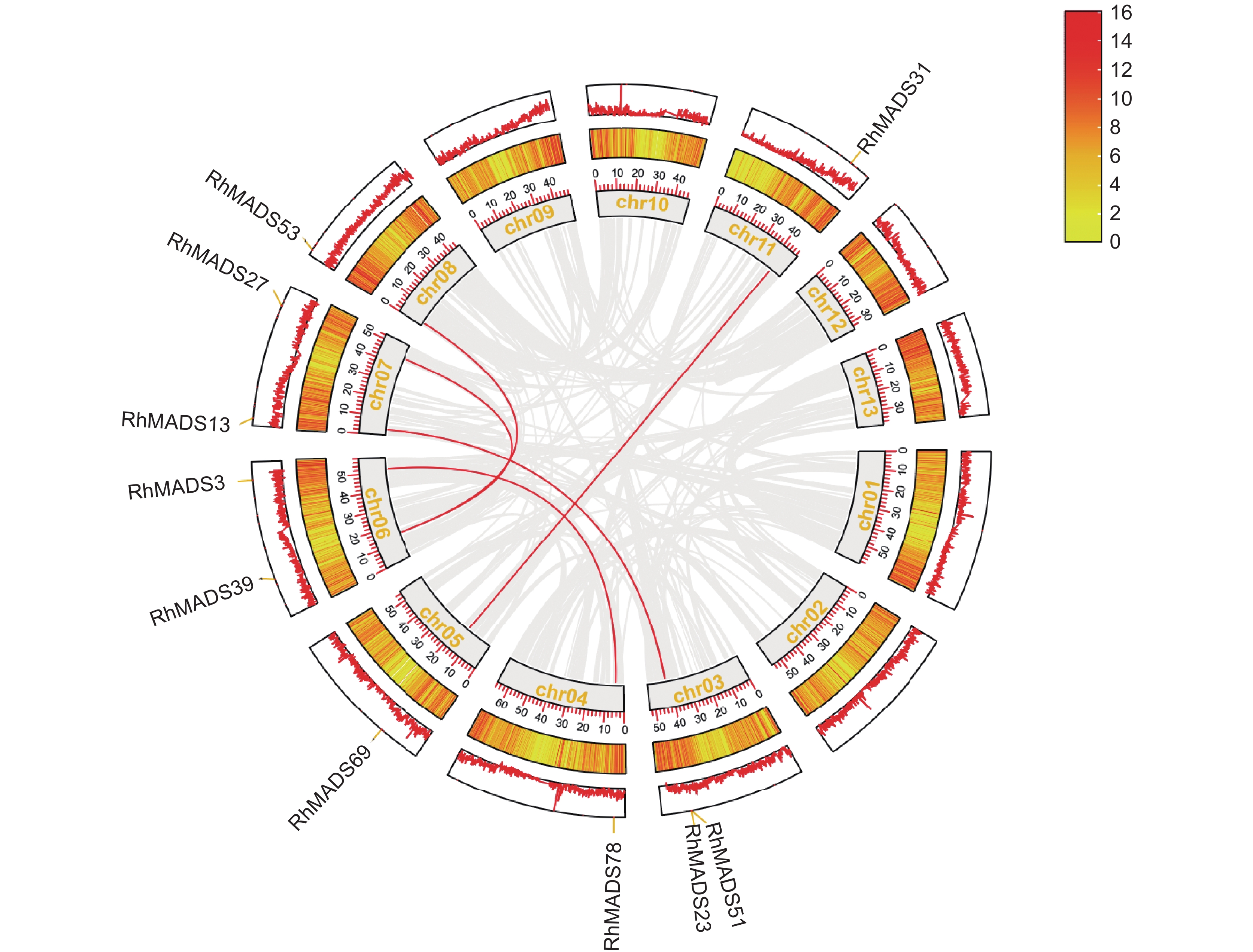
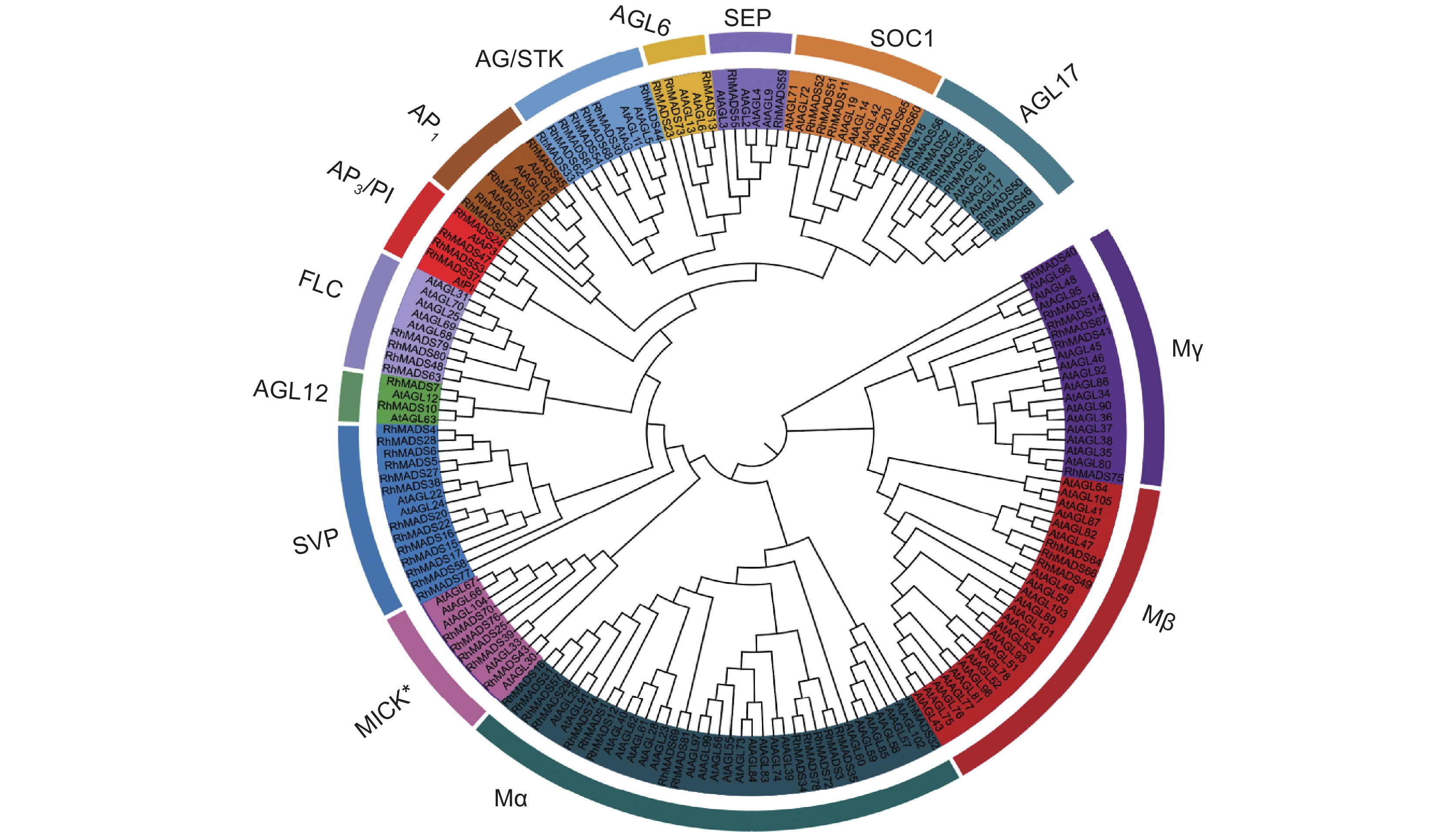
MADS-box是在植物生长发育过程中扮演重要角色的一类转录因子,特别是花器官形成、开花时间控制及果实发育与成熟等过程。本研究基于朱红大杜鹃(Rhododendron griersonianum Balf. f. et Forrest)全基因组测序数据,利用生物信息学方法从中鉴定出 81 个 MADS-box 基因,并进行了分析。根据系统发育关系和蛋白结构将 MADS-box分为两类:Type-Ⅰ型包含 24 个基因,Type-Ⅱ型包含57个基因,这些基因不均匀地分布于12条染色体上;81个MADS-box 基因中,存在 6 对片段复制和 1 对串联复制基因,且它们经历了纯化选择作用;同时,基因启动子区域含有光响应、植物生长、激素反应和胁迫响应等顺式作用元件。综上,本研究鉴定了朱红大杜鹃的MADS-box家族转录因子,为深入研究其 MADS-box 蛋白的生物学功能提供了基础。
-
关键词:
- 朱红大杜鹃 /
- MADS-box 基因家族 /
- 生物信息学 /
- 基因结构
Abstract:MADS-box genes play an important role in plant growth and development, especially in processes such as floral organ formation, flowering time regulation, and fruit development and ripening. Based on whole-genome sequencing data of Rhododendron griersonianum Balf. f. et Forrest, 81 MADS-box genes were identified and analyzed using bioinformatics methods. Phylogenetic analysis and protein structure classification divided these genes into two classes, including 24 genes in Type-Ⅰand 57 genes in Type-Ⅱ. The MADS-box genes were unevenly distributed across 12 chromosomes, with no genes located on chromosome 2. Among the 81 MADS-box genes, six pairs showed segmental duplications and one pair showed tandem duplication, all of which have undergone purifying selection. The promoter regions of the MADS-box genes contained elements involved in light response, plant growth, hormone response, and stress response. Overall, the identification of MADS-box gene family members provides a reliable reference for further studies on the biological functions of MADS-box proteins in R. griersonianum
-
Keywords:
- Rhododendron griersonianum /
- MADS-box gene family /
- Bioinformatics /
- Gene structure
-
1 1~2)如需查阅附件内容请登录《植物科学学报》网站(http://www.plantscience.cn)查看本期文章。2 3 4 1~4)如需查阅附件内容请登录《植物科学学报》网站(http://www.plantscience.cn)查看本期文章。5 1~2)如需查阅附件内容请登录《植物科学学报》网站(http://www.plantscience.cn)查看本期文章。6 -
-
[1] Liu DT,Sun WB,Ma YP,Fang ZD. Rediscovery and conservation of the critically endangered Rhododendron griersonianum in Yunnan,China[J]. Oryx,2019,53(1):14. doi: 10.1017/S0030605318001278
[2] Ma H,Liu YB,Liu DT,Sun WB,Liu XF,et al. Chromosome-level genome assembly and population genetic analysis of a critically endangered Rhododendron provide insights into its conservation[J]. Plant J,2021,107(5):1533−1545. doi: 10.1111/tpj.15399
[3] Gibbs D,Chamberlain D,Argent G. The Red List of Rhododendrons[M]. Richmond:Botanic Gardens Conservation International,2011:51.
[4] 覃海宁,杨永,董仕勇,何强,贾渝,等. 中国高等植物受威胁物种名录[J]. 生物多样性,2017,25(7):696−744. doi: 10.17520/biods.2017144 Qin HN,Yang Y,Dong SY,He Q,Jia Y,et al. Threatened species list of China’s higher plants[J]. Biodiversity Science,2017,25(7):696−744. doi: 10.17520/biods.2017144
[5] De Folter S,Angenent GC. Trans meets cis in MADS science[J]. Trends Plant Sci,2006,11(5):224−231. doi: 10.1016/j.tplants.2006.03.008
[6] Nam J,dePamphilis CW,Ma H,Nei M. Antiquity and evolution of the MADS-box gene family controlling flower development in plants[J]. Mol Biol Evol,2003,20(9):1435−1447. doi: 10.1093/molbev/msg152
[7] Theissen G,Becker A,Di Rosa A,Kanno A,Kim JT,et al. A short history of MADS-box genes in plants[J]. Plant Mol Biol,2000,42(1):115−149. doi: 10.1023/A:1006332105728
[8] Shan HY,Zhang N,Liu CJ,Xu GX,Zhang J,et al. Patterns of gene duplication and functional diversification during the evolution of the AP1/SQUA subfamily of plant MADS-box genes[J]. Mol Phylogenet Evol,2007,44(1):26−41. doi: 10.1016/j.ympev.2007.02.016
[9] Nam J,Kim J,Lee S,An G,Ma H,Nei M. TypeⅠMADS-box genes have experienced faster birth-and-death evolution than TypeⅡMADS-box genes in angiosperms[J]. Proc Natl Acad Sci USA,2004,101(7):1910−1915. doi: 10.1073/pnas.0308430100
[10] Alvarez-Buylla ER,Pelaz S,Liljegren SJ,Gold SE,Burgeff C,et al. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals[J]. Proc Natl Acad Sci USA,2000,97(10):5328−5333. doi: 10.1073/pnas.97.10.5328
[11] Par̆enicová L,de Folter S,Kieffer M,Horner DS,Favalli C,et al. Molecular and phylogenetic analyses of the complete MADS-Box transcription factor family in Arabidopsis:new openings to the MADS world[J]. Plant Cell,2003,15(7):1538−1551. doi: 10.1105/tpc.011544
[12] Qin Y,Zhu GP,Li FD,Wang L,Chen C,Zhao H. MIKC-Type MADS-box gene family discovery and evolutionary investigation in Rosaceae plants[J]. Agronomy,2023,13(7):1695. doi: 10.3390/agronomy13071695
[13] Henschel K,Kofuji R,Hasebe M,Saedler H,Münster T,Theißen G. Two ancient classes of MIKC-Type MADS-box genes are present in the moss Physcomitrella patens[J]. Mol Biol Evol,2002,19(6):801−814. doi: 10.1093/oxfordjournals.molbev.a004137
[14] Kwantes M,Liebsch D,Verelst W. How MIKC* MADS-box genes originated and evidence for their conserved function throughout the evolution of vascular plant gametophytes[J]. Mol Biol Evol,2012,29(1):293−302. doi: 10.1093/molbev/msr200
[15] Kofuji R,Sumikawa N,Yamasaki M,Kondo K,Ueda,K,et al. Evolution and divergence of the MADS-box gene family based on genome-wide expression analyses[J]. Mol Biol Evol,2003,20(12):1963−1977. doi: 10.1093/molbev/msg216
[16] Masiero S,Colombo L,Grini PE,Schnittger A,Kater MM. The emerging importance of TypeⅠMADS box transcription factors for plant reproduction[J]. Plant Cell,2011,23(3):865−872. doi: 10.1105/tpc.110.081737
[17] Alvarez-Buylla ER,Liljegren SJ,Pelaz S,Gold SE,Burgeff C,et al. MADS-box gene evolution beyond flowers:expression in pollen,endosperm,guard cells,roots and trichomes[J]. Plant Journal,2000,24(4):457−466.
[18] Chanderbali AS,Berger BA,Howarth DG,Soltis DE,Soltis PS. Evolution of floral diversity:genomics,genes and gamma[J]. Philos Trans R Soc B Biol Sci,2017,372(1713):20150509. doi: 10.1098/rstb.2015.0509
[19] Grimplet J,Martínez-Zapater JM,Carmona MJ. Structural and functional annotation of the MADS-box transcription factor family in grapevine[J]. BMC Genomics,2016,17(1):80. doi: 10.1186/s12864-016-2398-7
[20] Koo SC,Bracko O,Park MS,Schwab R,Chun HJ,et al. Control of lateral organ development and flowering time by the Arabidopsis thaliana MADS-box gene AGAMOUS-LIKE6[J]. Plant J,2010,62(5):807−816. doi: 10.1111/j.1365-313X.2010.04192.x
[21] Qiu YC,Köhler C,Sverlgel L. Endosperm evolution by duplicated and neofunctionalized TypeⅠMADS-box transcription factors[J]. Mol Biol Evol,2022,39(1):msab355. doi: 10.1093/molbev/msab355
[22] Schwarz-Sommer Z,Huijser P,Nacken W,Saedler H,Sommer H. Genetic control of flower development by homeotic genes in Antirrhinum majus[J]. Science,1990,250(4983):931−936. doi: 10.1126/science.250.4983.931
[23] Amasino RM,Michaels SD. The timing of flowering[J]. Plant Physiol,2010,154(2):516−520. doi: 10.1104/pp.110.161653
[24] Chen Q,Li J,Yang FJ. Genome-wide analysis of the MADS-box transcription factor family in Solanum melongena[J]. Int J Mol Sci,2023,24(1):826. doi: 10.3390/ijms24010826
[25] Arora R,Agarwal P,Ray S,Singh AK,Singh VP,et al. MADS-box gene family in rice:genome-wide identification,organization and expression profiling during reproductive development and stress[J]. BMC Genomics,2007,8(1):242. doi: 10.1186/1471-2164-8-242
[26] Wei B,Zhang RZ,Guo JJ,Liu DM,Li AL,et al. Genome-wide analysis of the MADS-box gene family in Brachypodium distachyon[J]. PLoS One,2014,9(1):e84781. doi: 10.1371/journal.pone.0084781
[27] Leseberg CH,Li AL,Kang H,Duvall M,Mao L. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa[J]. Gene,2006,378:84−94. doi: 10.1016/j.gene.2006.05.022
[28] Zhang XD,Fatima M,Zhou P,Ma Q,Ming R. Analysis of MADS-box genes revealed modified flowering gene network and diurnal expression in pineapple[J]. BMC Genomics,2020,21(1):8. doi: 10.1186/s12864-019-6421-7
[29] Ruelens P,Zhang ZC,van Mourik H,Maere S,Kaufmann K,Geuten K. The origin of floral organ identity quartets[J]. Plant Cell,2017,29(2):229−242. doi: 10.1105/tpc.16.00366
[30] Ferrario S,Immink RGH,Shchennikova A,Busscher-Lange J,Angenent GC. The MADS box gene FBP2 is required for SEPALLATA function in petunia[J]. Plant Cell,2003,15(4):914−925. doi: 10.1105/tpc.010280
[31] 高玮林,张力曼,薛超玲,张垚,刘孟军,赵锦. 枣E类MADS基因在花和果中的表达及其蛋白互作研究[J]. 园艺学报,2022,49(4):739−748. Gao WL,Zhang LM,Xue CL,Zhang Y,Liu MJ,Zhao J. Expression of E-Type MADS-box genes in flower and fruits and protein interaction analysis in Chinese jujube[J]. Acta Horticulturae Sinica,2022,49(4):739−748.
[32] 孙福辉,方慧仪,温小蕙,张亮生. 马银花MADS-box基因家族系统进化与表达分析[J]. 植物学报,2023,58(3):404−416. Sun FH,Fang HY,Wen XH,Zhang LS. Phylogenetic and expression analysis of MADS-box gene family in Rhododendron ovatum[J]. Chinese Bulletin of Botany,2023,58(3):404−416.
[33] Chen CJ,Chen H,Zhang Y,Thomas HR,Frank MH,et al. TBtools:an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant,2020,13(8):1194−1202. doi: 10.1016/j.molp.2020.06.009
[34] Zhang Z,Li J,Zhao XQ,Wang J,Wong GKS,Yu J. KaKs_calculator:calculating Ka and Ks through model selection and model averaging[J]. Genomics Proteomics Bioinformatics,2006,4(4):259−263. doi: 10.1016/S1672-0229(07)60007-2
[35] Shu YJ,Yu DS,Wang D,Guo DL,Guo CH. Genome-wide survey and expression analysis of the MADS-box gene family in soybean[J]. Mol Biol Rep,2013,40(6):3901−3911. doi: 10.1007/s11033-012-2438-6
[36] 潘志演,唐博希,田慧源,范吴蔚,彭剑涛,刘国琴. 烟草MADS-box基因家族鉴定及其腋芽发育相关基因表达分析[J]. 分子植物育种,2022,20(7):2217−2232. Pan ZY,Tang BX,Tian HY,Fan WW,Peng JT,Liu GQ. Identification of MADS-box gene family and expression analysis of genes related to axillary bud development in Nicotiana tabacum[J]. Molecular Plant Breeding,2022,20(7):2217−2232.
[37] Wang YS,Zhang JL,Hu ZL,Guo XH,Tian SB,Chen GP. Genome-wide analysis of the MADS-box transcription factor family in Solanum lycopersicum[J]. Int J Mol Sci,2019,20(12):2961. doi: 10.3390/ijms20122961
[38] Li C,Wang Y,Xu L,Nie SS,Chen YL,et al. Genome-wide characterization of the MADS-box gene family in radish (Raphanus sativus L.) and assessment of its roles in flowering and floral organogenesis[J]. Front Plant Sci,2016,7:1390.
[39] Won SY,Jung JA,Kim JS. Genome-wide analysis of the MADS-box gene family in Chrysanthemum[J]. Comput Biol Chem,2021,90:107424. doi: 10.1016/j.compbiolchem.2020.107424
[40] 高欢,郑珂昕,廖光联,王海令,陈璐,等. 中华猕猴桃全基因组MADS-box基因家族鉴定及表达分析[J]. 果树学报,2023,40(11):2307−2324. Gao H,Zheng KX,Liao GL,Wang HL,Chen L,et al. Genome-wide identification and expression analysis of the MADS-box gene family in Actinidia chinensis[J]. Journal of Fruit Science,2023,40(11):2307−2324.
[41] Wei X,Wang LH,Yu JY,Zhang YX,Li DH,Zhang XR. Genome-wide identification and analysis of the MADS-box gene family in sesame[J]. Gene,2015,569(1):66−76. doi: 10.1016/j.gene.2015.05.018
[42] 王溪唯,陈璨,王大玮. 云南栘[木衣]MADS-box基因家族鉴定与表达分析[J]. 生物工程学报,2023,39(7):2897−2913. Wang XW,Chen C,Wang DW. Identification and expression analysis of MADS-box gene family in Docynia delavayi (Franch.) Schneid[J]. Chinese Journal of Biotechnology,2023,39(7):2897−2913.
[43] Li D,Liu C,Shen LS,Wu Y,Chen HY,et al. A repressor complex governs the integration of flowering signals in Arabidopsis[J]. Dev Cell,2008,15(1):110−120. doi: 10.1016/j.devcel.2008.05.002
[44] Yang YF,Zhu T,Niu DK. Association of intron loss with high mutation rate in Arabidopsis:implications for genome size evolution[J]. Genome Biol Evol,2013,5(4):723−733. doi: 10.1093/gbe/evt043
[45] Wu JY,Xiao JF,Wang LP,Zhong J,Yin HY,et al. Systematic analysis of intron size and abundance parameters in diverse lineages[J]. Sci China Life Sci,2013,56(10):968−974. doi: 10.1007/s11427-013-4540-y
[46] Gao HH,Wang ZM,Li SL,Hou ML,Zhou Y,et al. Genome-wide survey of potato MADS-box genes reveals that StMADS1 and StMADS13 are putative downstream targets of tuberigen StSP6A[J]. BMC Genomics,2018,19(1):726. doi: 10.1186/s12864-018-5113-z
[47] Airoldi CA,Davies B. Gene duplication and the evolution of plant MADS-box transcription factors[J]. J Genet Genomics,2012,39(4):157−165. doi: 10.1016/j.jgg.2012.02.008
[48] Zhou PY,Qu YS,Wang ZW,Huang B,Wen Q,et al. Gene structural specificity and expression of MADS-box gene family in Camellia chekiangoleosa[J]. Int J Mol Sci,2023,24(4):3434. doi: 10.3390/ijms24043434
-
其他相关附件
-
PDF格式
柳文-附件 点击下载(1390KB)
-



 下载:
下载: