Genome-wide identification and expression analysis of the MAP70 gene family in Brassica napus L.
-
摘要:
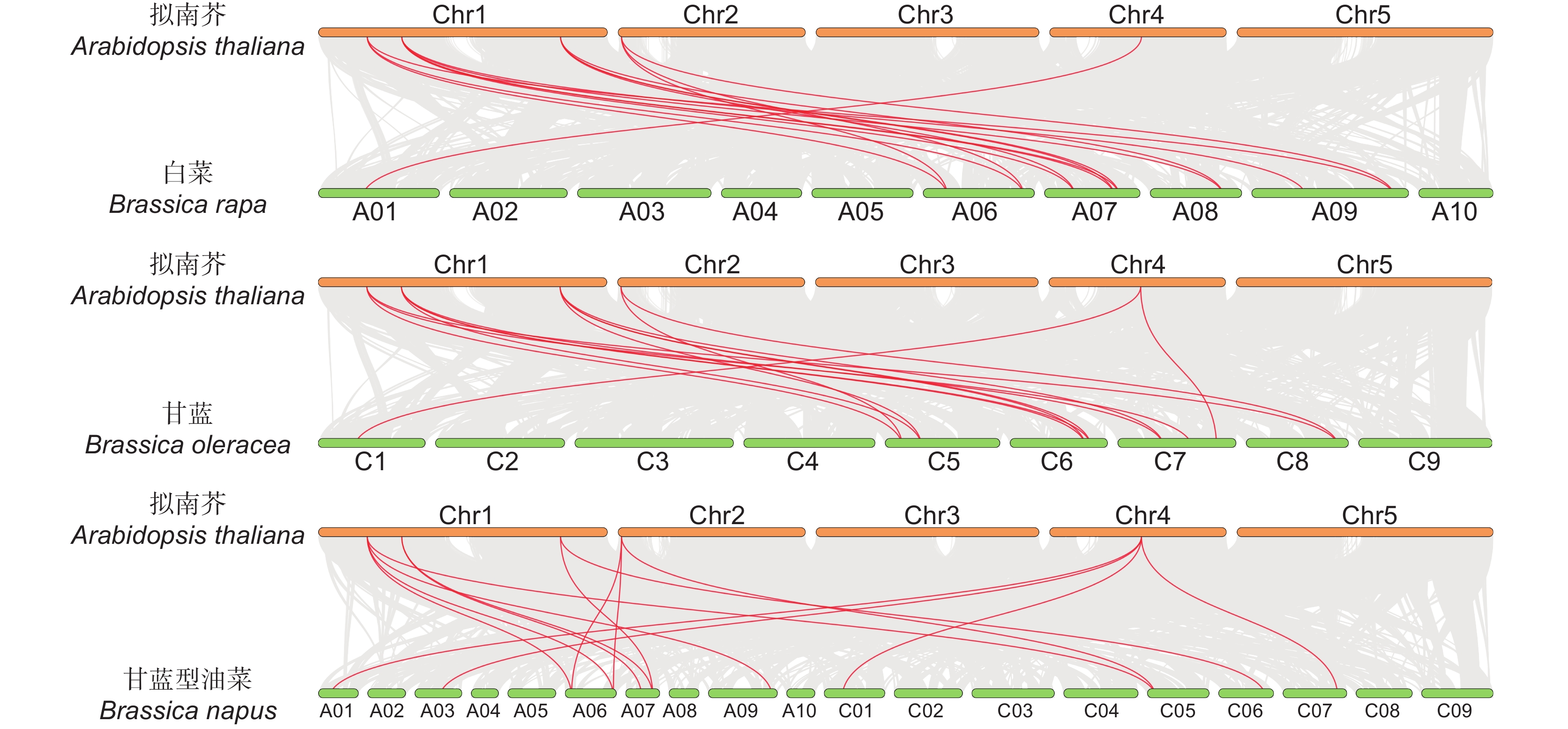
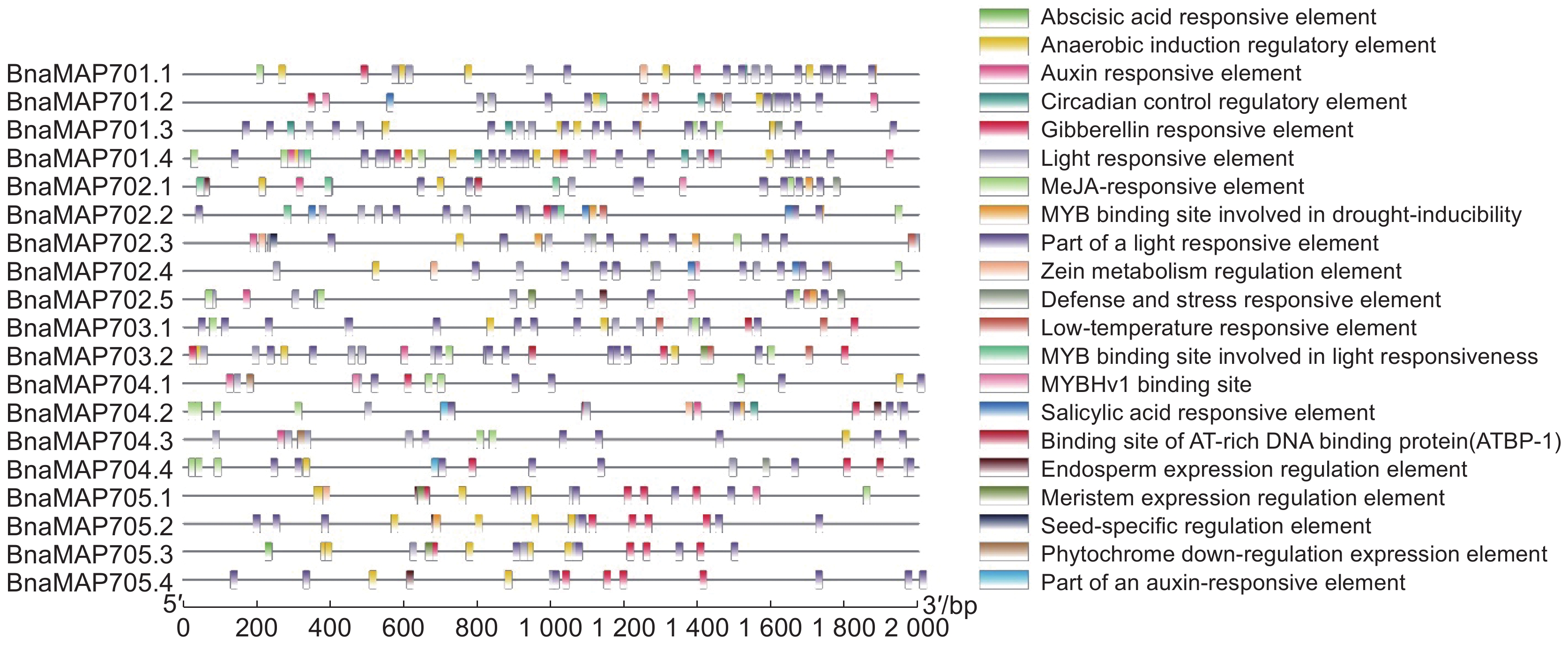
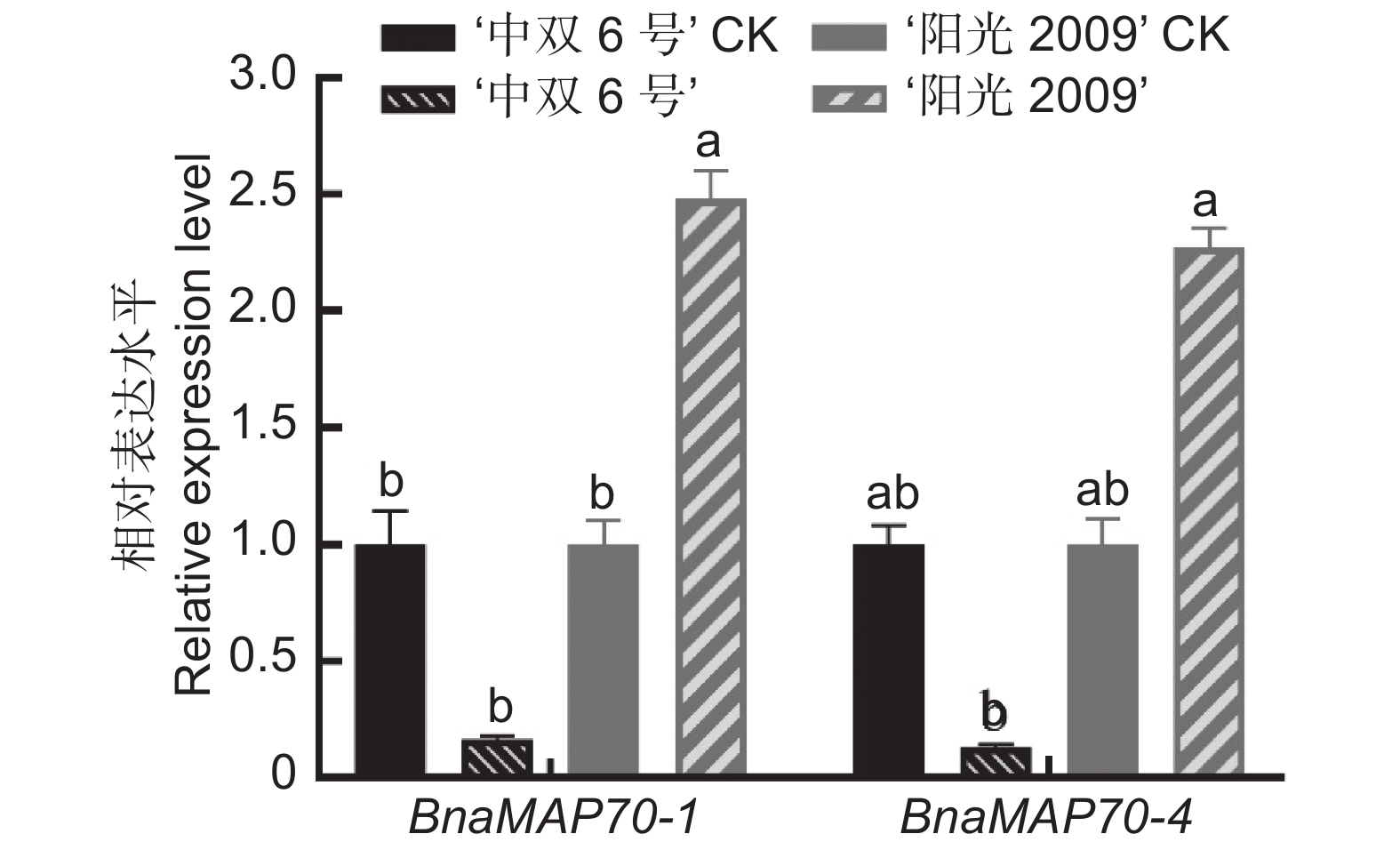
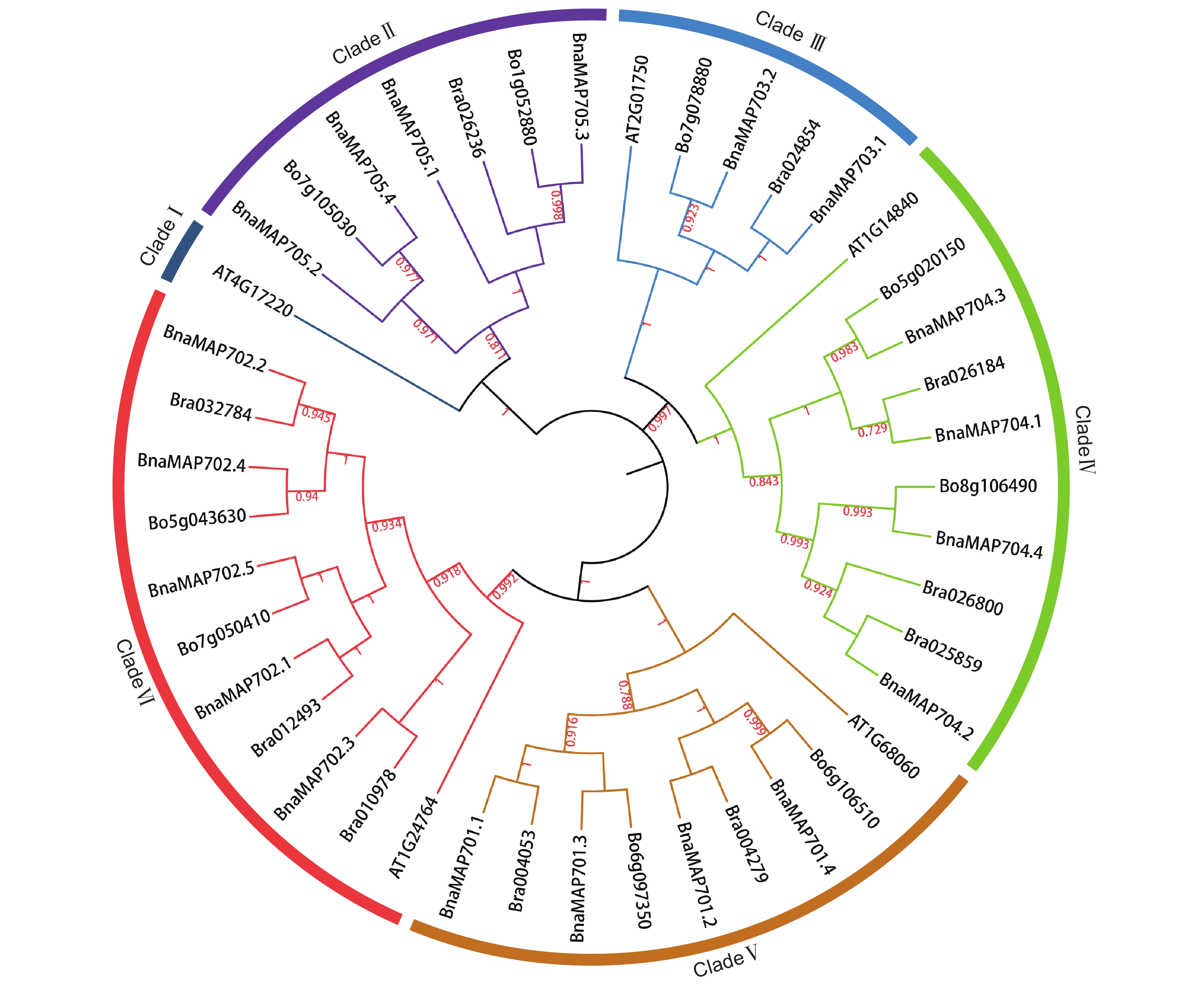
采用生物信息学方法,从甘蓝型油菜(Brassica napus L.)基因组数据库中筛选鉴定MAP70s基因家族成员,对鉴定得到的BnaMAP70s基因家族成员的序列特征进行生物信息学分析,同时利用qRT-PCR方法分析该基因家族成员在不耐渍和强耐渍甘蓝型油菜品种幼苗中的基因表达水平。结果显示,本研究共鉴定得到19个BnaMAP70基因家族成员,分布在11条染色体上,可分为5个亚家族。BnaMAP70s启动子的上游存在厌氧胁迫、响应植物激素等相关元件,表明BnaMAP70s可能参与植株生长发育和渍水胁迫调控;转录水平存在品种特异性、植株部位特异性、时间特异性。qRT-PCR分析结果表明,BnaMAP70-1和BnaMAP70-4在渍水胁迫下出现表达差异,说明这两个基因受渍水胁迫的调控。
-
关键词:
- 甘蓝型油菜 /
- BnaMAP70s基因家族 /
- 耐渍 /
- 生物信息学
Abstract:MAP70 family members were screened and identified using bioinformatics based on the genomic database of Brassica napus L., with bioinformatics analysis performed on the sequence characteristics of the identified BnaMAP70 gene family members. Expression levels of the BnaMAP70 genes were analyzed using qRT-PCR in seedling varieties with waterlogging sensitivity and tolerance. In total, 19 members of the BnaMAP70 gene family were identified, distributed on 11 chromosomes and divided into five subfamilies. The upstream elements involved in anaerobic stress and phytohormones of the BnaMAP70 promoter indicated that they may be involved in plant growth, development, and regulation of waterlogging stress. The transcription levels of genes were species-specific, plant site-specific, and time-specific. The qRT-PCR results showed that BnaMAP70-1 and BnaMAP70-4 were differentially expressed under waterlogging stress, indicating that these two genes may be regulated by waterlogging stress.
-
Keywords:
- Brassica napus /
- BnaMAP70 gene family /
- Waterlogging tolerance /
- Bioinformatics
-
1 1~ 3)如需查阅附件内容请登录《植物科学学报》网站(http://www.plantscience.cn)查看本期文章。2 1 ~ 3)如需查阅附件内容请登录《植物科学学报》网站(http://www.plantscience.cn)查看本期文章。 -
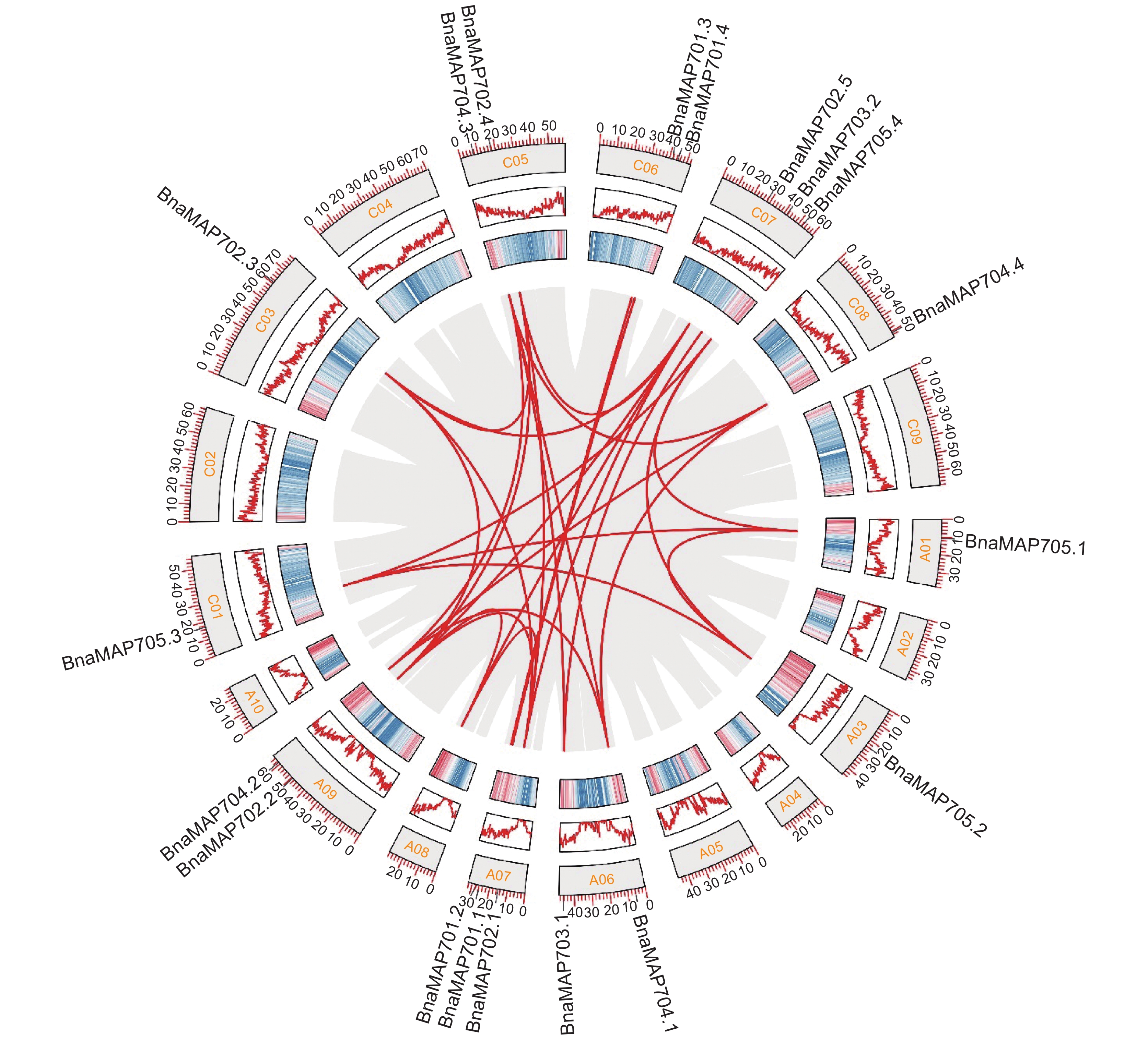
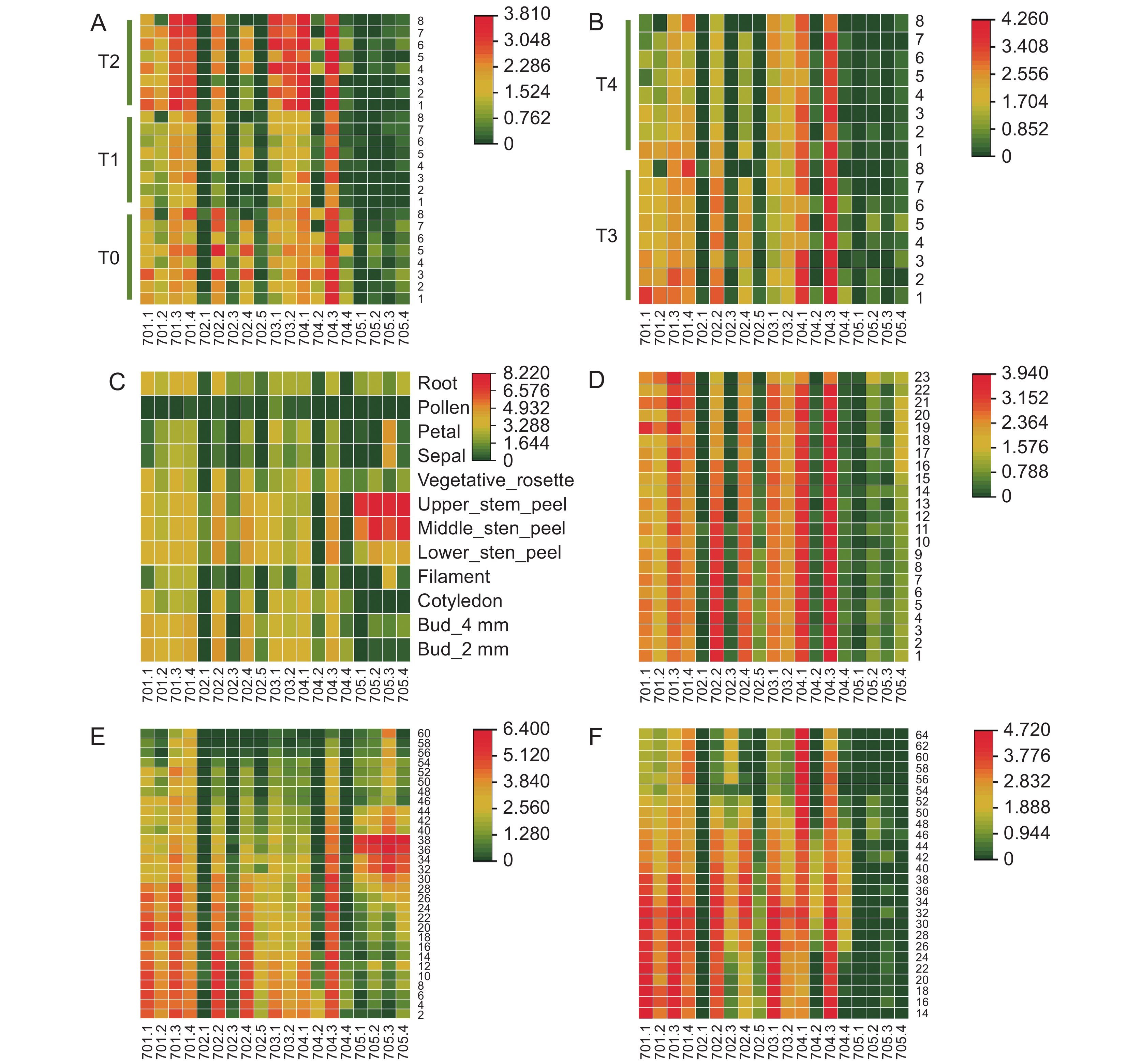
图 5 BnaMAP70s转录水平分析
701.1~705.4 分别对应基因 BnaMAP701.1~BnaMAP705.4。A、B:不同甘蓝型油菜 (1:‘Quinta’;2:‘Tapidor’;3:‘Shengli’;4:‘Zheyou’;5:‘Gangan’;6:‘ZS11’;7:‘Westar’;8:‘NO2127’) 叶片中 BnaMAP70s 基因转录水平 (T0~T5分别为播种后第24、54、82、115、147 天);C:BnaMAP70s 在 ‘ZS11’ 不同部位的基因转录水平;D:BnaMAP70s 在 ‘ZS11’ 不同时期叶片中的基因转录水平 (1~23为第1~23片叶);E:BnaMAP70s 在 ‘ZS11’ 不同时期角果中的基因转录水平 (2~60为开花后第2~60 天);F:BnaMAP70s 在 ‘ZS11’ 不同时期种子中的基因转录水平 (14~64为开花后第 14~64 天)。
Figure 5. Transcriptional levels of BnaMAP70s
701.1-705.4 in the heat map correspond to the genes BnaMAP701.1-BnaMAP705.4 respectively. A,B: Transcription levels of BnaMAP70s gene in leaves (T0~T5: 24, 54, 82, 115, 147 day after sowing) of different B. napus (1: ‘Quinta’; 2: ‘Tapidor’; 3: ‘Shengli’; 4: ‘Zheyou’; 5: ‘Gangan’; 6: ‘ZS11’; 7: ‘Westar’; 8: ‘NO2127’); C: Gene transcription levels of BnaMAP70s in different parts of ‘ZS11’; D: Gene transcription levels of BnaMAP70s in leaves at different periods of ‘ZS11’(1~23: leaf 1-23); E: Gene transcription levels of BnaMAP70s in silique at different periods of ‘ZS11’(2~60: 2-60 day after flowering); F: Gene transcription levels of BnaMAP70s in seed at different periods of ‘ZS11’(14~64: 14-64 day after flowering).
-
[1] Fan XL,Zhang ZS,Gao HY,Yang C,Liu MJ,et al. Photoinhibition-like damage to the photosynthetic apparatus in plant leaves induced by submergence treatment in the dark[J]. PLoS One,2014,9 (2):e89067. doi: 10.1371/journal.pone.0089067
[2] Sundgren TK,Uhlen AK,Lillemo M,Briese C,Wojciechowski T. Rapid seedling establishment and a narrow root stele promotes waterlogging tolerance in spring wheat[J]. J Plant Physiol,2018,227:45−55. doi: 10.1016/j.jplph.2018.04.010
[3] Lynch JP. Root phenes that reduce the metabolic costs of soil exploration:opportunities for 21st century agriculture[J]. Plant Cell Environ,2015,38 (9):1775−1784. doi: 10.1111/pce.12451
[4] Ploschuk RA,Miralles DJ,Colmer TD,Ploschuk EL,Striker GG. Waterlogging of winter crops at early and late stages:impacts on leaf physiology,growth and yield[J]. Front Plant Sci,2018,9:1863. doi: 10.3389/fpls.2018.01863
[5] Ambros S,Kotewitsch M,Wittig PR,Bammer B,Mustroph A. Transcriptional response of two Brassica napus cultivars to short-term hypoxia in the root zone[J]. Front Plant Sci,2022,13:897673. doi: 10.3389/fpls.2022.897673
[6] Yamauchi T,Colmer TD,Pedersen O,Nakazono M. Regulation of root traits for internal aeration and tolerance to soil waterlogging-flooding stress[J]. Plant Physiol,2018,176 (2):1118−1130. doi: 10.1104/pp.17.01157
[7] Sauter M. Root responses to flooding[J]. Curr Opin Plant Biol,2013,16 (3):282−286. doi: 10.1016/j.pbi.2013.03.013
[8] Gibbs DJ,Lee SC,Isa N,Gramuglia S,Fukao T,et al. Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants[J]. Nature,2011,479 (7373):415−418. doi: 10.1038/nature10534
[9] Tong C,Hill CB,Zhou GF,Zhang XQ,Jia Y,Li CD. Opportunities for improving waterlogging tolerance in cereal crops-physiological traits and genetic mechanisms[J]. Plants (Basel)
,2021,10 (8):1560. doi: 10.3390/plants10081560 [10] 马海清,刘清云,高立兵,胡志文,张清香,等. 油菜初花期淹水胁迫对产量及构成因子的影响[J]. 中国农业文摘-农业工程,2020,32(6):77−80. doi: 10.3969/j.issn.1002-5103.2020.06.029 [11] 俞建河,王本来,曹秀清,沈涛. 淹水时期与天数对油菜生长性状和产量的影响[J]. 安徽农业科学,2021,49(14):184−187. doi: 10.3969/j.issn.0517-6611.2021.14.049 Yu JH,Wang BL,Cao XQ,Shen T. Effects of flooding period and days on the growth traits and yield of rapeseed[J]. Journal of Anhui Agricultural Sciences,2021,49 (14):184−187. doi: 10.3969/j.issn.0517-6611.2021.14.049
[12] 李阳阳,荆蓉蓉,吕蓉蓉,石鹏程,李欣,等. 甘蓝型油菜湿害胁迫响应性状的全基因组关联分析及候选基因预测[J]. 作物学报,2019,45(12):1806−1821. Li YY,Jing RR,LÜ RR,Shi PC,Li X,et al. Genome-wide association analysis and candidate genes prediction of waterlogging-responding traits in Brassica napus L.[J]. Acta Agronomica Sinica,2019,45 (12):1806−1821.
[13] 徐明月. 甘蓝型油菜发芽期耐渍相关基因的筛选[D]. 北京: 中国农业科学院, 2014: 31. [14] Zou XL,Zeng L,Lu GY,Cheng Y,Xu JS,Zhang XK. Comparison of transcriptomes undergoing waterlogging at the seedling stage between tolerant and sensitive varieties of Brassica napus L.[J]. J Integr Agr,2015,14 (9):1723−1734. doi: 10.1016/S2095-3119(15)61138-8
[15] Zou XL,Tan XY,Hu CW,Zeng L,Lu GP,et al. The transcriptome of Brassica napus L. roots under waterlogging at the seedling stage[J]. Int J Mol Sci,2013,14 (2):2637−2651. doi: 10.3390/ijms14022637
[16] Li JJ,Iqbal S,Zhang YT,Chen YH,Tan ZD,et al. Transcriptome analysis reveals genes of flooding-tolerant and flooding-sensitive rapeseeds differentially respond to flooding at the germination stage[J]. Plants (Basel)
,2021,10 (4):693. doi: 10.3390/plants10040693 [17] 李继军. 油菜响应苗期渍害的动态过程和遗传基础[D]. 武汉: 华中农业大学, 2021: 17. [18] Poole RL. The TAIR database[J]. Methods Mol Biol,2007,406:179−212.
[19] Song JM,Liu DX,Xie WZ,Yang ZQ,Guo L,et al. BnPIR:Brassica napus pan-genome information resource for 1689 accessions[J]. Plant Biotechnol J,2021,19 (3):412−414. doi: 10.1111/pbi.13491
[20] Chen CJ,Chen H,Zhang Y,Thomas HR,Frank MH,et al. TBtools:an integrative toolkit developed for interactive analyses of big biological data[J]. Mol Plant,2020,13 (8):1194−1202. doi: 10.1016/j.molp.2020.06.009
[21] Mistry J,Chuguransky S,Williams L,Qureshi M,Salazar GA,et al. Pfam:The protein families database in 2021[J]. Nucleic Acids Res,2021,49 (D1):D412−D419. doi: 10.1093/nar/gkaa913
[22] McWilliam H,Li WZ,Uludag M,Squizzato S,Park YM,et al. Analysis tool web services from the EMBL-EBI[J]. Nucleic Acids Res,2013,41:W597−W600. doi: 10.1093/nar/gkt376
[23] Johnson M,Zaretskaya I,Raytselis Y,Merezhuk Y,McGinnis S,Madden TL. NCBI BLAST:a better web interface[J]. Nucleic Acids Res,2008,36 (W):W5−W9.
[24] Yang MZ,Derbyshire MK,Yamashita RA,Marchler-Bauer A. NCBI's conserved domain database and tools for protein domain analysis[J]. Curr Protoc Bioinformatics,2020,69 (1):e90.
[25] Gasteiger E,Gattiker A,Hoogland C,Ivanyi I,Appel RD,Bairoch A. ExPASy:the proteomics server for in-depth protein knowledge and analysis[J]. Nucleic Acids Res,2003,31 (13):3784−3788. doi: 10.1093/nar/gkg563
[26] Horton P,Park KJ,Obayashi T,Fujita N,Harada H,et al. WoLF PSORT:protein localization predictor[J]. Nucleic Acids Res,2007,35 (W):W585−W587.
[27] Bolser DM, Staines DM, Perry E, Kersey PJ. Ensembl plants: integrating tools for visualizing, mining, and analyzing plant genomic data[M]//Van Dijk ADJ, ed. Plant Genomics Databases. New York: Humana Press, 2017, 1533: 1-31.
[28] Hall BG. Building phylogenetic trees from molecular data with MEGA[J]. Mol Biol Evol,2013,30 (5):1229−1235. doi: 10.1093/molbev/mst012
[29] Letunic I,Bork P. Interactive Tree Of Life (iTOL) v5:an online tool for phylogenetic tree display and annotation[J]. Nucleic Acids Res,2021,49 (W1):W293−W296. doi: 10.1093/nar/gkab301
[30] Hu B,Jin JP,Guo AY,Zhang H,Luo JC,Gao G. GSDS 2.0:an upgraded gene feature visualization server[J]. Bioinformatics,2015,31 (8):1296−1297. doi: 10.1093/bioinformatics/btu817
[31] Bailey TL,Boden M,Buske FA,Frith M,Grant CE,et al. MEME SUITE:tools for motif discovery and searching[J]. Nucleic Acids Res,2009,37 (W):W202−W208.
[32] Lescot M,Déhais P,Thijs G,Marchal K,Moreau Y,et al. PlantCARE,a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences[J]. Nucleic Acids Res,2002,30 (1):325−327. doi: 10.1093/nar/30.1.325
[33] Liu DX,Yu LQ,Wei LL,Yu PG,Wang J,et al. BnTIR:an online transcriptome platform for exploring RNA-seq libraries for oil crop Brassica napus[J]. Plant Biotechnol J,2021,19 (10):1895−1897. doi: 10.1111/pbi.13665
[34] Livak KJ,Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the
2−ΔΔCT Method[J]. Methods,2001,25 (4):402−408. doi: 10.1006/meth.2001.1262[35] Lee YRJ,Liu B. Microtubule nucleation for the assembly of acentrosomal microtubule arrays in plant cells[J]. New Phytol,2019,222 (4):1705−1718. doi: 10.1111/nph.15705
[36] Goodson HV,Jonasson EM. Microtubules and microtubule-associated proteins[J]. Cold Spring Harb Perspect Biol,2018,10 (6):a022608. doi: 10.1101/cshperspect.a022608
[37] Struk S,Dhonukshe P. MAPs:cellular navigators for microtubule array orientations in Arabidopsis[J]. Plant Cell Rep,2014,33 (1):1−21. doi: 10.1007/s00299-013-1486-2
[38] Brouhard GJ,Rice LM. The contribution of αβ-tubulin curvature to microtubule dynamics[J]. J Cell Biol,2014,207 (3):323−334. doi: 10.1083/jcb.201407095
[39] Gardiner J. The evolution and diversification of plant microtubule-associated proteins[J]. Plant J,2013,75 (2):219−229. doi: 10.1111/tpj.12189
[40] Korolev AV,Chan J,Naldrett MJ,Doonan JH,Lloyd CW. Identification of a novel family of 70 kDa microtubule-associated proteins in Arabidopsis cells[J]. Plant J,2005,42 (4):547−555. doi: 10.1111/j.1365-313X.2005.02393.x
[41] Panchy N,Lehti-Shiu M,Shiu SH. Evolution of gene duplication in plants[J]. Plant Physiol,2016,171 (4):2294−2316. doi: 10.1104/pp.16.00523
[42] Zaman MS,Malik AI,Kaur P,Erskine W. Waterlogging tolerance of pea at germination[J]. J Agro Crop Sci,2018,204 (2):155−164. doi: 10.1111/jac.12230
[43] Bao ZR, Guo Y, Deng YL, Zang JZ, Zhang JH, et al. The microtubule-associated protein SlMAP70 interacts with SlIQD21 and regulates fruit shape formation in tomato[J/OL]. BioRxiv, 2022. doi: 10.1101/2022.08.08.503161.
[44] Stöckle D,Reyes-Hernández BJ,Barro AV,Nenadić M,Winter Z,et al. Microtubule-based perception of mechanical conflicts controls plant organ morphogenesis[J]. Sci Adv,2022,8 (6):eabm4974. doi: 10.1126/sciadv.abm4974
[45] Arduini I,Kokubun M,Licausi F. Editorial:crop response to waterlogging[J]. Front Plant Sci,2019,10:1578. doi: 10.3389/fpls.2019.01578
[46] Liu MM,Tan XF,Sun XH,Zwiazek JJ. Properties of root water transport in canola (Brassica napus) subjected to waterlogging at the seedling,flowering and podding growth stages[J]. Plant and Soil,2020,454 (1):431−445.
[47] Ryser P,Gill HK,Byrne CJ. Constraints of root response to waterlogging in Alisma triviale[J]. Plant Soil,2011,343 (1):247−260.
[48] Pesquet E,Korolev AV,Calder G,Lloyd CW. Mechanisms for shaping,orienting,positioning and patterning plant secondary cell walls[J]. Plant Signal Behav,2011,6 (6):843−849. doi: 10.4161/psb.6.6.15202
[49] Pesquet E,Korolev AV,Calder G,Lloyd CW. The microtubule-associated protein AtMAP70-5 regulates secondary wall patterning in Arabidopsis wood cells[J]. Curr Biol,2010,20 (8):744−749. doi: 10.1016/j.cub.2010.02.057
-
期刊类型引用(2)
1. 谢伶俐,李永铃,许本波,张学昆. 油菜耐渍机理解析及遗传改良研究进展. 作物学报. 2025(02): 287-300 .  百度学术
百度学术
2. 蒋晓晗,聂笑一. 甘蓝型油菜基因组缺失变异检测研究. 粮油与饲料科技. 2024(06): 199-201 .  百度学术
百度学术
其他类型引用(0)
-
其他相关附件
-
PDF格式
黄郢附件 点击下载(1015KB)
-



 下载:
下载: